













| General Information: |
|
| Name(s) found: |
UTP13 /
YLR222C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 817 amino acids |
Gene Ontology: |
|
| Cellular Component: |
small nucleolar ribonucleoprotein complex
[IPI |
| Biological Process: |
maturation of SSU-rRNA
[IMP |
| Molecular Function: |
snoRNA binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLKTSYKGI SLNPIYAGSS AVATVSENGK ILATPVLDEI NIIDLTPGSR KILHKISNED 60
61 EQEITALKLT PDGQYLTYVS QAQLLKIFHL KTGKVVRSMK ISSPSYILDA DSTSTLLAVG 120
121 GTDGSIIVVD IENGYITHSF KGHGGTISSL KFYGQLNSKI WLLASGDTNG MVKVWDLVKR 180
181 KCLHTLQEHT SAVRGLDIIE VPDNDEPSLN LLSGGRDDII NLWDFNMKKK CKLLKTLPVN 240
241 QQVESCGFLK DGDGKRIIYT AGGDAIFQLI DSESGSVLKR TNKPIEELFI IGVLPILSNS 300
301 QMFLVLSDQT LQLINVEEDL KNDEDTIQVT SSIAGNHGII ADMRYVGPEL NKLALATNSP 360
361 SLRIIPVPDL SGPEASLPLD VEIYEGHEDL LNSLDATEDG LWIATASKDN TAIVWRYNEN 420
421 SCKFDIYAKY IGHSAAVTAV GLPNIVSKGY PEFLLTASND LTIKKWIIPK PTASMDVQII 480
481 KVSEYTRHAH EKDINALSVS PNDSIFATAS YDKTCKIWNL ENGELEATLA NHKRGLWDVS 540
541 FCQYDKLLAT SSGDKTVKIW SLDTFSVMKT LEGHTNAVQR CSFINKQKQL ISCGADGLIK 600
601 IWDCSSGECL KTLDGHNNRL WALSTMNDGD MIVSADADGV FQFWKDCTEQ EIEEEQEKAK 660
661 LQVEQEQSLQ NYMSKGDWTN AFLLAMTLDH PMRLFNVLKR ALGESRSRQD TEEGKIEVIF 720
721 NEELDQAISI LNDEQLILLM KRCRDWNTNA KTHTIAQRTI RCILMHHNIA KLSEIPGMVK 780
781 IVDAIIPYTQ RHFTRVDNLV EQSYILDYAL VEMDKLF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
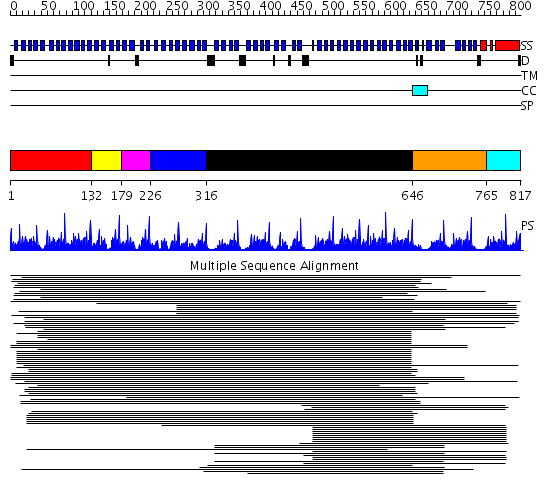
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..131] | 3.75 | Tup1, C-terminal domain | |
| 2 | View Details | [132..178] | 5.420216 | WD domain, G-beta repeat No confident structure predictions are available. | |
| 3 | View Details | [179..225] | 5.142668 | WD domain, G-beta repeat No confident structure predictions are available. | |
| 4 | View Details | [226..315] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [316..645] | 179.758605 | beta1-subunit of the signal-transducing G protein heterotrimer | |
| 6 | View Details | [646..764] | 74.218487 | Groucho/tle1, C-terminal domain | |
| 7 | View Details | [765..817] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)