













| General Information: |
|
| Name(s) found: |
BMS1 /
YPL217C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1183 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA mitochondrion [IDA nuclear outer membrane [IDA nucleolus [IDA |
| Biological Process: |
ribosome assembly
[IDA rRNA processing [IDA |
| Molecular Function: |
GTP binding
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEQSNKQHRK AKEKNTAKKK LHTQGHNAKA FAVAAPGKMA RTMQRSSDVN ERKLHVPMVD 60
61 RTPEDDPPPF IVAVVGPPGT GKTTLIRSLV RRMTKSTLND IQGPITVVSG KHRRLTFLEC 120
121 PADDLNAMID IAKIADLVLL LIDGNFGFEM ETMEFLNIAQ HHGMPRVLGV ATHLDLFKSQ 180
181 STLRASKKRL KHRFWTEVYQ GAKLFYLSGV INGRYPDREI LNLSRFISVM KFRPLKWRNE 240
241 HPYMLADRFT DLTHPELIET QGLQIDRKVA IYGYLHGTPL PSAPGTRVHI AGVGDFSVAQ 300
301 IEKLPDPCPT PFYQQKLDDF EREKMKEEAK ANGEITTAST TRRRKRLDDK DKLIYAPMSD 360
361 VGGVLMDKDA VYIDIGKKNE EPSFVPGQER GEGEKLMTGL QSVEQSIAEK FDGVGLQLFS 420
421 NGTELHEVAD HEGMDVESGE ESIEDDEGKS KGRTSLRKPR IYGKPVQEED ADIDNLPSDE 480
481 EPYTNDDDVQ DSEPRMVEID FNNTGEQGAE KLALETDSEF EESEDEFSWE RTAANKLKKT 540
541 ESKKRTWNIG KLIYMDNISP EECIRRWRGE DDDSKDESDI EEDVDDDFFR KKDGTVTKEG 600
601 NKDHAVDLEK FVPYFDTFEK LAKKWKSVDA IKERFLGAGI LGNDNKTKSD SNEGGEELYG 660
661 DFEDLEDGNP SEQAEDNSDK ESEDEDENED TNGDDDNSFT NFDAEEKKDL TMEQEREMNA 720
721 AKKEKLRAQF EIEEGENFKE DDENNEYDTW YELQKAKISK QLEINNIEYQ EMTPEQRQRI 780
781 EGFKAGSYVR IVFEKVPMEF VKNFNPKFPI VMGGLLPTEI KFGIVKARLR RHRWHKKILK 840
841 TNDPLVLSLG WRRFQTLPIY TTTDSRTRTR MLKYTPEHTY CNAAFYGPLC SPNTPFCGVQ 900
901 IVANSDTGNG FRIAATGIVE EIDVNIEIVK KLKLVGFPYK IFKNTAFIKD MFSSAMEVAR 960
961 FEGAQIKTVS GIRGEIKRAL SKPEGHYRAA FEDKILMSDI VILRSWYPVR VKKFYNPVTS1020
1021 LLLKEKTEWK GLRLTGQIRA AMNLETPSNP DSAYHKIERV ERHFNGLKVP KAVQKELPFK1080
1081 SQIHQMKPQK KKTYMAKRAV VLGGDEKKAR SFIQKVLTIS KAKDSKRKEQ KASQRKERLK1140
1141 KLAKMEEEKS QRDKEKKKEY FAQNGKRTTM GGDDESRPRK MRR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
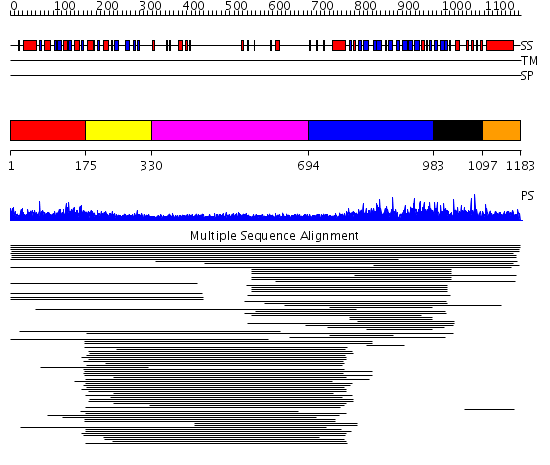
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..174] | 2.253997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [175..329] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [330..693] | 2.221849 | Heavy meromyosin subfragment | |
| 4 | View Details | [694..982] | 2.429937 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [983..1096] | 1.027985 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [1097..1183] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)