













| General Information: |
|
| Name(s) found: |
GCN1 /
YGL195W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2672 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA cytosol [IDA |
| Biological Process: |
regulation of translational elongation
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTAILNWEDI SPVLEKGTRE SHVSKRVPFL QDISQLVRQE TLEKPQLSEI AFVLLNTFTI 60
61 YEDNRSKSLV TSILLDILNL EPCLLENFIR FISDVVISNP ATKAVADYLN LLDWINSFLI 120
121 FVSHNSNLFE EYIPKLLVAH SYATFGVETI LDNQEEGKKS QDKQNQHRKR IRYCIFQTTV 180
181 KAFLKCLKDN DDSISFMKIS IKTVLESYSK LKITSVGVVM IMGALTQAAL QLLSRQPALH 240
241 SVLKENSAEK YCEYLGKEVF LGKNPPSSFC LEIGLKPFLK EFVSQELFIK FFIPNIEKAV 300
301 LRSPEVGFSI LSELYAGVSP EKVNLLNAFA SSKLINQYFS SFKSSKEVVR SVSLQSMIIL 360
361 LRKISNTDTT LEDLTKLIDE IFKNIKSNLN ADYKSLISKI LIEIPLTHYE VSEKICKGLS 420
421 PYIGKEGNEA ALTLMLNAFF VHYFSLGKPI EDLDKIISAG FADKKPALKK CWFAAFLNNS 480
481 NAASEEVILN FIDGCLEFVK DSIIHYQTHG HACILASIEF TNKILALDNT ELNDRVMQLI 540
541 ETLPENSSIG DAILTAALST ELSIENRIHA VNLLQELFYK KPEFIGFSVI DAIERRMRVQ 600
601 ELIPQQNTSF KYVTSVLLAI TSELPDKEAS IKVLINALVI AQWNIFNIKN GWAGLVLRAR 660
661 LDPAEVVKEH ASVIMEKILE ITGSCEWIDT IYGACGLQAA AYAAFIQPNE FTPILCKTIE 720
721 ADLTADDFSR LSEEDFEIFA GEEGVLVVDV LEESMNKKLS NKNSKEYETL MWEQKIRKEQ 780
781 AKKNVKKLSK EEQELVNEQL AKESAVRSHV SEISTRLKRG IRLVSELSKA ACLVQNGIAT 840
841 WFPLAVTKLL YLCSEPNISK LTEDVNNVFL QLSQNVSERL GNIRLFLGLA TLRVHNANGI 900
901 SQDYLQEPLV ELLTRVLFRI KFVSNQAAID SISLTYILPL LINVLEKGKA IALKNADKPV 960
961 VKAEFVEEDE EEEHLLLAME IISVHAEAFE DPSIPRISIV EVLLSLLSLP SKAKIAKDCF1020
1021 NALCQSISVA PNQEDLDMIL SNLLSPNQFV RSTILETLDN EFELEPFMKY SPEVFICRFD1080
1081 SDPSNREIAD FIWEFNKFVV NDELLKSLFP LFNQDDSGLR LFAANAYAFG AVSLFTSEEN1140
1141 SSKDYLNDLL NFYKEKAKPL EPILDQFGLV LVSASEQKDP WQGRSTVAIT LKIMAKAFSA1200
1201 EDDTVVNIIK FLVDDGGLVD REPIVRQEMK EAGVELITLH GSQNSKDLIP IFEEALSSST1260
1261 DSALKENVII LYGTLARHLQ QSDARIHTII ERLLSTLDTP SADIQQAVSA CIAPLVFQFK1320
1321 QKVGDYLGIL MEKLLNPTVA SSMRKGAAWG IAGLVKGYGI SALSEFDIIR NLIEAAEDKK1380
1381 EPKRRESVGF CFQYLSESLG KFFEPYVIEI LPNILKNLGD AVPEVRDATA RATKAIMAHT1440
1441 TGYGVKKLIP VAVSNLDEIA WRTKRGSVQL LGNMAYLDPT QLSASLSTIV PEIVGVLNDS1500
1501 HKEVRKAADE SLKRFGEVIR NPEIQKLVPV LLQAIGDPTK YTEEALDSLI QTQFVHYIDG1560
1561 PSLALIIHII HRGMHDRSAN IKRKACKIVG NMAILVDTKD LIPYLQQLID EVEIAMVDPV1620
1621 PNTRATAARA LGALVERLGE EQFPDLIPRL LDTLSDESKS GDRLGSAQAL AEVISGLGLT1680
1681 KLDEMLPTIL AGVTNFRAYI REGFMPLLLF LPVCFGSQFA PYINQIIQPI LSGLADNDEN1740
1741 IRDTALKAGK LIVKNYATKA VDLLLPELER GMFDENDRIR LSSVQLTGEL LFQVTGISSR1800
1801 NEFSEEDGDH NGEFSGKLVD VLGQDRRDRI LAALFVCRND TSGIVRATTV DIWKALVPNT1860
1861 PRAVKEILPT LTGMIVTHLA SSSNVLRNIA AQTLGDLVRR VGGNALSQLL PSLEESLIET1920
1921 SNSDSRQGVC IALYELIESA STETISQFQS TIVNIIRTAL IDESATVREA AALSFDVFQD1980
1981 VVGKTAVDEV LPYLLHMLES SDNSDFALLG LQEIMSKKSD VIFPILIPTL LAPPIDAFRA2040
2041 SALGSLAEVA GSALYKRLSI IINALVDAII GTSEDESTKG ALELALDRVF LSVNDDEGLH2100
2101 PLLQQIMSLL KSDNIEKRIA VLERLPNFFD KTVLDFDVYI PNFVSHAILS LDDEDQRVVN2160
2161 GNFNALSTLL KKVDKPTLEK LVKPAKQSLA LTGRQGQDVA AFKLPRGPNC VLPIFLHGLM2220
2221 YGSNDEREES ALAIADVVSK TPAANLKPFV SVITGPLIRV VGERFSSDIK AAILFALNVL2280
2281 FIKIPMFLRP FIPQLQRTFV KSLSDATNET LRLRAAKALG ALIEHQPRVD PLVIELVTGA2340
2341 KQATDEGVKT AMLKALLEVI MKAGSKLNEN SKTNIVNLVE EEMLGSNDKL AVAYAKLIGS2400
2401 LSEILSNDEA HKILQDKVLN ADLDGETGKF AILTLNSFLK DAPTHIFNTG LIDEFVSYIL2460
2461 NAIRSPDVYF GENGTIAAGK LLLLEGEKRS PFVKKDAAEP FKIGDENINL LINELSKAVL2520
2521 QPASNSTDVR RLALVVIRTL ARFKFDECIK QYFDVVGPSV FSCLRDPVIP IKLAAEKAYL2580
2581 ALFKLVEEDD MHTFNEWFAK ISDRGNSIET VTGTTIQLRS VGDYTKRVGK RLANVERERI2640
2641 AAGGDAETMF SDRFEDEREI WAVGGVELTT DI |
The following runs contain data for this protein:
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
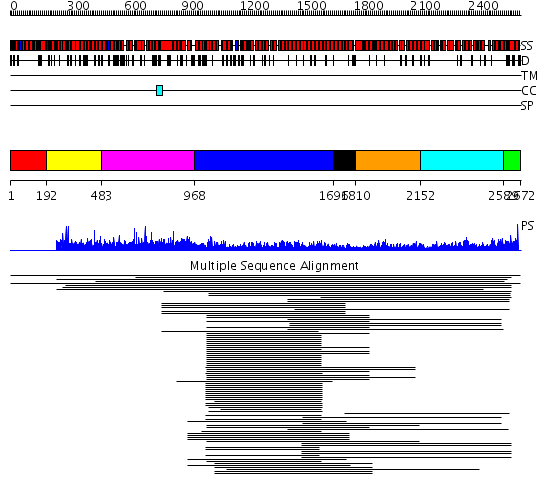
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..191] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [192..482] | 2.007993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [483..967] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [968..1695] | 19.461998 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1696..1809] | N/A | Confident ab initio structure predictions are available. | |
| 6 | View Details | [1810..2151] | 4.253988 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [2152..2588] | 8.169996 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [2589..2672] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.36 |
Source: Reynolds et al. (2008)