













| General Information: |
|
| Name(s) found: |
SAM1 /
YLR180W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 382 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
methionine metabolic process
[TAS |
| Molecular Function: |
methionine adenosyltransferase activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGTFLFTSE SVGEGHPDKI CDQVSDAILD ACLAEDPHSK VACETAAKTG MIMVFGEITT 60
61 KAQLDYQKIV RDTIKKIGYD DSAKGFDYKT CNVLVAIEQQ SPDIAQGVHE EKDLEDIGAG 120
121 DQGIMFGYAT DETPEGLPLT ILLAHKLNMA MADARRDGSL AWLRPDTKTQ VTVEYKDDHG 180
181 RWVPQRIDTV VVSAQHADEI TTEDLRAQLK SEIIEKVIPR DMLDENTKYF IQPSGRFVIG 240
241 GPQGDAGLTG RKIIVDAYGG ASSVGGGAFS GKDYSKVDRS AAYAARWVAK SLVAAGLCKR 300
301 VQVQFSYAIG IAEPLSLHVD TYGTATKSDE EIIDIISKNF DLRPGVLVKE LDLARPIYLP 360
361 TASYGHFTNQ EYPWEKPKTL KF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Green EM, et al (2005) | |
| View Run | 3906: TAP-tagged | Green EM, et al (2005) | |
| View Run | 3908: TAP-tagged, includes hir2(delta) in background | Green EM, et al (2005) | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
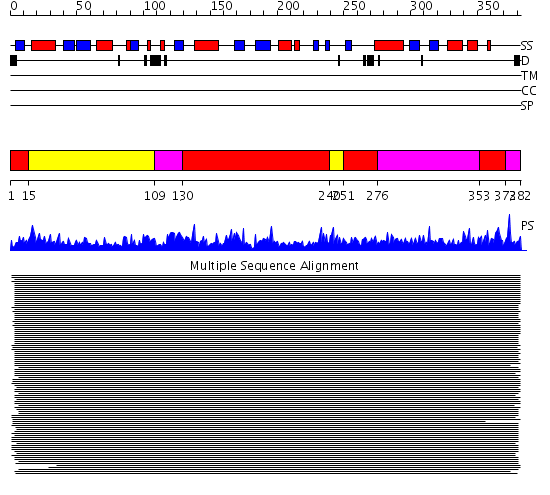
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..14] [130..239] [251..275] [353..371] |
2570.0 | S-adenosylmethionine synthetase | |
| 2 | View Details | [15..108] [240..250] |
2570.0 | S-adenosylmethionine synthetase | |
| 3 | View Details | [109..129] [276..352] [372..382] |
2570.0 | S-adenosylmethionine synthetase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)