













| General Information: |
|
| Name(s) found: |
UTP9 /
YHR196W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 575 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nucleolus [IDA small nucleolar ribonucleoprotein complex [IPI |
| Biological Process: |
maturation of SSU-rRNA
[IMP ribosome biogenesis [RCA |
| Molecular Function: |
snoRNA binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSSLDLVAS FSHDSTRFAF QASVAQKNNV DIYPLNETKD YVVNSSLVSH IDYETNDMKV 60
61 SDVIFFGWCS DLIDTQSSNI KRKLDEDEGT GESSEQRCEN FFVNGFPDGR IVVYSSNGKD 120
121 IVNIIKNKKE ILGADTDESD IWILDSDKVV KKLQYNNSKP LKTFTLVDGK DDEIVHFQIL 180
181 HQNGTLLVCI ITKQMVYIVD PSKRRPSTKY SFEISDAVAC EFSSDGKYLL IANNEELIAY 240
241 DLKEDSKLIQ SWPVQVKTLK TLDDLIMALT TDGKINNYKI GEADKVCSIV VNEDLEIIDF 300
301 TPINSKQQVL ISWLNVNEPN FESISLKEIE TQGYITINKN EKNNADEADQ KKLEEKEEEA 360
361 QPEVQHEKKE TETKINKKVS KSDQVEIANI LSSHLEANST EILDDLMSGS WTEPEIKKFI 420
421 LTKINTVDHL SKIFLTISKS ITQNPWNEEN LLPLWLKWLL TLKSGELNSI KDKHTKKNCK 480
481 HLKSALRSSE EILPVLLGIQ GRLEMLRRQA KLREDLAQLS MQEGEDDEIE VIEHSNVISN 540
541 PLQDQASPVE KLEPDSIVYA NGESDEFVDA SEYKD |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Unpublished Data |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
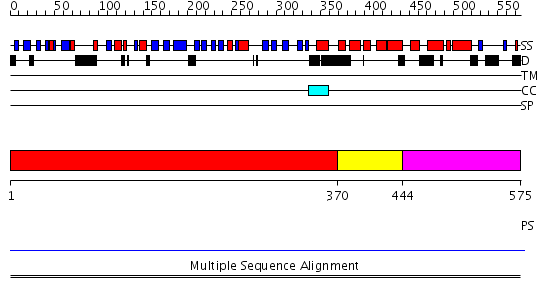
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..369] | 1.54 | beta1-subunit of the signal-transducing G protein heterotrimer | |
| 2 | View Details | [370..443] | 1.64 | Tup1, C-terminal domain | |
| 3 | View Details | [444..575] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)