













| General Information: |
|
| Name(s) found: |
UTP4 /
YDR324C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 776 amino acids |
Gene Ontology: |
|
| Cellular Component: |
small nucleolar ribonucleoprotein complex
[IPI |
| Biological Process: |
maturation of SSU-rRNA
[IMP ribosome biogenesis [RCA |
| Molecular Function: |
snoRNA binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSLLSVLK EKSRSLKIRN KPVKMTSQER MIVHRCRFVD FTPATITSLA FSHKSNINKL 60
61 TPSDLRLAIG RSNGNIEIWN PRNNWFQEMV IEGGKDRSIE GLCWSNVNGE SLRLFSIGGS 120
121 TVVTEWDLAT GLPLRNYDCN SGVIWSISIN DSQDKLSVGC DNGTVVLIDI SGGPGVLEHD 180
181 TILMRQEARV LTLAWKKDDF VIGGCSDGRI RIWSAQKNDE NMGRLLHTMK VDKAKKESTL 240
241 VWSVIYLPRT DQIASGDSTG SIKFWDFQFA TLNQSFKAHD ADVLCLTTDT DNNYVFSAGV 300
301 DRKIFQFSQN TNKSQKNNRW VNSSNRLLHG NDIRAICAYQ SKGADFLVSG GVEKTLVINS 360
361 LTSFSNGNYR KMPTVEPYSK NVLVNKEQRL VVSWSESTVK IWTMGTDSST EQNYKLVCKL 420
421 TLKDDQNIST CSLSPDGQVL VVGRPSTTKV FHLQPVGNKL KVTKLDNDLL LRTSTKLVKF 480
481 IDNSKIVICS CEDDVFIVDL ESEEDEKPQE VELLEVTSTK SSIKVPYINR INHLEVDQNI 540
541 AVISRGCGVV DILDLKARIS KPLARLNNFI TAVHINTSRK SVVVITADNK IYEFNMNLNS 600
601 EAENEDSESV LTQWSKNNTD NLPKEWKTLK ENCVGIFSDI ENSSRLWFWG ATWISRIDFD 660
661 VDFPINKRRK QKKRTHEGLT ITDESNFMND EEDDEDDDID MEISENLNVL LNQGNKIKST 720
721 DVQRNEESSG HFFFTDKYKP LLFVDLISSN ELAIIERNPL TFHSKQKAFI QPKLVF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Unpublished Data |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
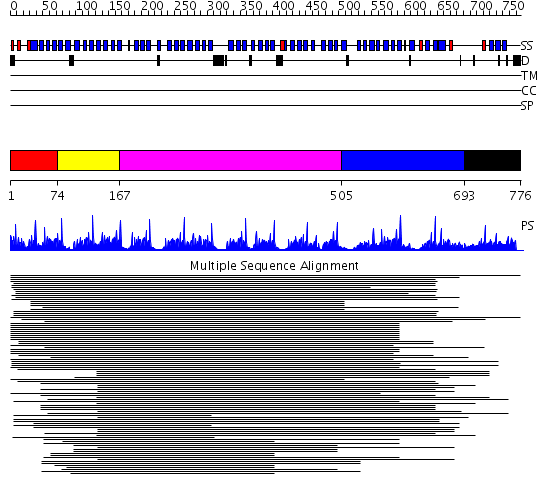
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..73] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [74..166] | 79.30103 | beta1-subunit of the signal-transducing G protein heterotrimer | |
| 3 | View Details | [167..504] | 96.24799 | Tup1, C-terminal domain | |
| 4 | View Details | [505..692] | 3.485995 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [693..776] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)