













| General Information: |
|
| Name(s) found: |
CKB2 /
YOR039W
[SGD]
|
| Description(s) found:
Found 36 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 258 amino acids |
Gene Ontology: |
|
| Cellular Component: |
protein kinase CK2 complex
[IDA |
| Biological Process: |
G2/M transition of mitotic cell cycle
[IPI G1/S transition of mitotic cell cycle [IPI flocculation via cell wall protein-carbohydrate interaction [IMP protein amino acid phosphorylation [IDA response to DNA damage stimulus [IDA regulation of transcription from RNA polymerase III promoter [IDA regulation of transcription from RNA polymerase I promoter [IDA cellular ion homeostasis [IMP |
| Molecular Function: |
protein kinase CK2 regulator activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSRSENVGT VTREGSRVEQ DDVLMDDDSD SSEYVDMWID LFLGRKGHEY FCDVDPEYIT 60
61 DRFNLMNLQK TVSKFSYVVQ YIVDDLDDSI LENMTHARLE QLESDSRKLY GLIHARYIIT 120
121 IKGLQKMYAK YKEADFGRCP RVYCNLQQLL PVGLHDIPGI DCVKLYCPSC EDLYIPKSSR 180
181 HSSIDGAYFG TSFPGMFLQA FPDMVPKHPT KRYVPKIFGF ELHKQAQLTR WQELQRLKLV 240
241 EKLESKDVDL TKSGGFKT |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
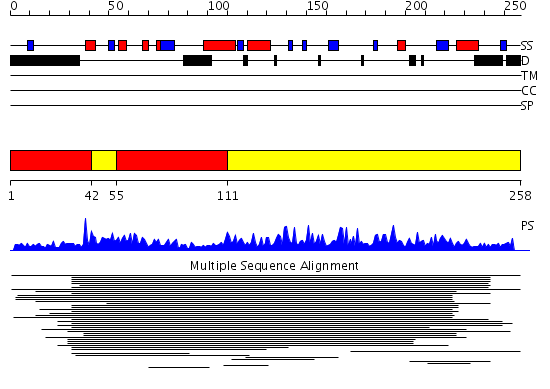
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..41] [55..110] |
630.0 | Casein kinase II beta subunit | |
| 2 | View Details | [42..54] [111..258] |
630.0 | Casein kinase II beta subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)