













| General Information: |
|
| Name(s) found: |
NHP10 /
YDL002C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 203 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA Ino80 complex [IPI |
| Biological Process: |
chromatin remodeling
[IPI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVEEKKRRL EELKDQNVVL GLAIQRSRLS VKRLKLEYGV LLERLESRIE LDPELNCEDP 60
61 LPTLASFKQE LLTKPFRKSK TKRHKVKERD PNMPKRPTNA YLLYCEMNKE RIRQNGSLDV 120
121 TRDLAEGWKN LNEQDRKPYY KLYSEDRERY QMEMEIYNKK ISNIDADDDK EENEQKIKNN 180
181 EEGSSTKVAD SKGGEDGSLV SSN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ACT1 ARP4 ARP5 ARP8 IES1 IES2 IES3 IES5 INO80 NHP10 RVB1 RVB2 TAF14 |
| View Details | Krogan NJ, et al. (2006) |

|
ARP4 ARP5 ARP6 ARP8 CCA1 EAF1 EAF5 EAF6 EAF7 EPL1 ESA1 IES1 IES2 IES3 IES4 IES5 IES6 INO80 NHP10 NTO1 PIH1 RVB1 RVB2 SAS3 SWC3 SWC4 SWC5 SWC7 SWR1 TAH1 TRA1 VPS71 VPS72 YAF9 YNG2 |
| View Details | Gavin AC, et al. (2006) |
|
ARP5 ARP8 IES1 IES3 IES5 INO80 NHP10 |
| View Details | Gavin AC, et al. (2002) |

|
ABF2 ACT1 ADH1 ARP4 ARP8 CKA1 CKA2 CKB1 CKB2 CTI6 DEP1 DOT6 EAF3 ECM16 ESC8 HTA2 IES1 IES5 INO80 IOC3 ISW1 ISW2 ITC1 MED4 MOT1 NHP10 NPL6 NRD1 PHO23 RCO1 RPC40 RPD3 RSC2 RSC4 RSC58 RSC6 RSC8 RVB2 RXT2 RXT3 SAP30 SDS3 SIN3 STO1 TOP1 TUP1 UME1 UME6 VPS1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
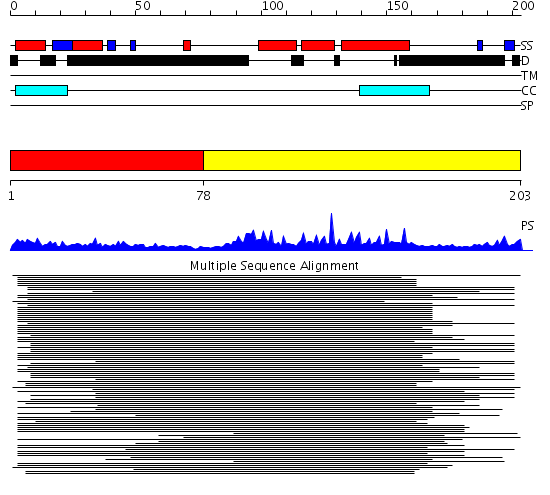
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | 8.045757 | HMG1, domains A and B | |
| 2 | View Details | [78..203] | 73.69897 | NHP6a |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)