













| General Information: |
|
| Name(s) found: |
TRA1 /
YHR099W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 3744 amino acids |
Gene Ontology: |
|
| Cellular Component: |
NuA4 histone acetyltransferase complex
[IPI SLIK (SAGA-like) complex [IPI SAGA complex [IDA histone acetyltransferase complex [IPI |
| Biological Process: |
DNA repair
[IDA histone acetylation [IDA regulation of transcription from RNA polymerase II promoter [IPI |
| Molecular Function: |
histone acetyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLTEQIEQF ASRFRDDDAT LQSRYSTLSE LYDIMELLNS PEDYHFFLQA VIPLLLNQLK 60
61 EVPISYDAHS PEQKLRNSML DIFNRCLMNQ TFQPYAMEVL EFLLSVLPKE NEENGILCMK 120
121 VLTTLFKSFK SILQDKLDSF IRIIIQIYKN TPNLINQTFY EAGKAEQGDL DSPKEPQADE 180
181 LLDEFSKNDE EKDFPSKQSS TEPRFENSTS SNGLRSSMFS FKILSECPIT MVTLYSSYKQ 240
241 LTSTSLPEFT PLIMNLLNIQ IKQQQEAREQ AESRGEHFTS ISTEIINRPA YCDFILAQIK 300
301 ATSFLAYVFI RGYAPEFLQD YVNFVPDLII RLLQDCPSEL SSARKELLHA TRHILSTNYK 360
361 KLFLPKLDYL FDERILIGNG FTMHETLRPL AYSTVADFIH NIRSELQLSE IEKTIKIYTG 420
421 YLLDESLALT VQIMSAKLLL NLVERILKLG KENPQEAPRA KKLLMIIIDS YMNRFKTLNR 480
481 QYDTIMKYYG RYETHKKEKA EKLKNSIQDN DKESEEFMRK VLEPSDDDHL MPQPKKEDIN 540
541 DSPDVEMTES DKVVKNDVEM FDIKNYAPIL LLPTPTNDPI KDAFYLYRTL MSFLKTIIHD 600
601 LKVFNPPPNE YTVANPKLWA SVSRVFSYEE VIVFKDLFHE CIIGLKFFKD HNEKLSPETT 660
661 KKHFDISMPS LPVSATKDAR ELMDYLAFMF MQMDNATFNE IIEQELPFVY ERMLEDSGLL 720
721 HVAQSFLTSE ITSPNFAGIL LRFLKGKLKD LGNVDFNTSN VLIRLFKLSF MSVNLFPNIN 780
781 EVVLLPHLND LILNSLKYST TAEEPLVYFY LIRTLFRSIG GGRFENLYRS IKPILQVLLQ 840
841 SLNQMILTAR LPHERELYVE LCITVPVRLS VLAPYLPFLM KPLVFALQQY PDLVSQGLRT 900
901 LELCIDNLTA EYFDPIIEPV IDDVSKALFN LLQPQPFNHA ISHNVVRILG KLGGRNRQFL 960
961 KPPTDLTEKT ELDIDAIADF KINGMPEDVP LSVTPGIQSA LNILQSYKSD IHYRKSAYKY1020
1021 LTCVLLLMTK SSAEFPTNYT ELLKTAVNSI KLERIGIEKN FDLEPTVNKR DYSNQENLFL1080
1081 RLLESVFYAT SIKELKDDAM DLLNNLLDHF CLLQVNTTLL NKRNYNGTFN IDLKNPNFML1140
1141 DSSLILDAIP FALSYYIPEV REVGVLAYKR IYEKSCLIYG EELALSHSFI PELAKQFIHL1200
1201 CYDETYYNKR GGVLGIKVLI DNVKSSSVFL KKYQYNLANG LLFVLKDTQS EAPSAITDSA1260
1261 EKLLIDLLSI TFADVKEEDL GNKVLENTLT DIVCELSNAN PKVRNACQKS LHTISNLTGI1320
1321 PIVKLMDHSK QFLLSPIFAK PLRALPFTMQ IGNVDAITFC LSLPNTFLTF NEELFRLLQE1380
1381 SIVLADAEDE SLSTNIQKTT EYSTSEQLVQ LRIACIKLLA IALKNEEFAT AQQGNIRIRI1440
1441 LAVFFKTMLK TSPEIINTTY EALKGSLAEN SKLPKELLQN GLKPLLMNLS DHQKLTVPGL1500
1501 DALSKLLELL IAYFKVEIGR KLLDHLTAWC RVEVLDTLFG QDLAEQMPTK IIVSIINIFH1560
1561 LLPPQADMFL NDLLLKVMLL ERKLRLQLDS PFRTPLARYL NRFHNPVTEY FKKNMTLRQL1620
1621 VLFMCNIVQR PEAKELAEDF EKELDNFYDF YISNIPKNQV RVVSFFTNMV DLFNTMVITN1680
1681 GDEWLKKKGN MILKLKDMLN LTLKTIKENS FYIDHLQLNQ SIAKFQALYL RFTELSERDQ1740
1741 NPLLLDFIDF SFSNGIKASY SLKKFIFHNI IASSNKEKQN NFINDATLFV LSDKCLDARI1800
1801 FVLKNVINST LIYEVATSGS LKSYLVEDKK PKWLELLHNK IWKNSNAILA YDVLDHHDLF1860
1861 RFELLQLSAI FIKADPEIIA EIKKDIIKFC WNFIKLEDTL IKQSAYLVTS YFISKFDFPI1920
1921 KVVTQVFVAL LRSSHVEARY LVKQSLDVLT PVLHERMNAA GTPDTWINWV KRVMVENSSS1980
1981 QNNILYQFLI SHPDLFFNSR DLFISNIIHH MNKITFMSNS NSDSHTLAID LASLILYWEN2040
2041 KTLEITNVNN TKTDSDGDVV MSDSKSDINP VEADTTAIIV DANNNSPISL HLREACTAFL2100
2101 IRYVCASNHR AIETELGLRA INILSELISD KHWTNVNVKL VYFEKFLIFQ DLDSENILYY2160
2161 CMNALDVLYV FFKNKTKEWI MENLPTIQNL LEKCIKSDHH DVQEALQKVL QVIMKAIKAQ2220
2221 GVSVIIEEES PGKTFIQMLT SVITQDLQET SSVTAGVTLA WVLFMNFPDN IVPLLTPLMK2280
2281 TFSKLCKDHL SISQPKDAMA LEEARITTKL LEKVLYILSL KVSLLGDSRR PFLSTVALLI2340
2341 DHSMDQNFLR KIVNMSRSWI FNTEIFPTVK EKAAILTKML AFEIRGEPSL SKLFYEIVLK2400
2401 LFDQEHFNNT EITVRMEQPF LVGTRVEDIG IRKRFMTILD NSLERDIKER LYYVIRDQNW2460
2461 EFIADYPWLN QALQLLYGSF NREKELSLKN IYCLSPPSIL QEYLPENAEM VTEVNDLELS2520
2521 NFVKGHIASM QGLCRIISSD FIDSLIEIFY QDPKAIHRAW VTLFPQVYKS IPKNEKYGFV2580
2581 RSIITLLSKP YHTRQISSRT NVINMLLDSI SKIESLELPP HLVKYLAISY NAWYQSINIL2640
2641 ESIQSNTSID NTKIIEANED ALLELYVNLQ EEDMFYGLWR RRAKYTETNI GLSYEQIGLW2700
2701 DKAQQLYEVA QVKARSGALP YSQSEYALWE DNWIQCAEKL QHWDVLTELA KHEGFTDLLL2760
2761 ECGWRVADWN SDRDALEQSV KSVMDVPTPR RQMFKTFLAL QNFAESRKGD QEVRKLCDEG2820
2821 IQLSLIKWVS LPIRYTPAHK WLLHGFQQYM EFLEATQIYA NLHTTTVQNL DSKAQEIKRI2880
2881 LQAWRDRLPN TWDDVNMWND LVTWRQHAFQ VINNAYLPLI PALQQSNSNS NINTHAYRGY2940
2941 HEIAWVINRF AHVARKHNMP DVCISQLARI YTLPNIEIQE AFLKLREQAK CHYQNMNELT3000
3001 TGLDVISNTN LVYFGTVQKA EFFTLKGMFL SKLRAYEEAN QAFATAVQID LNLAKAWAQW3060
3061 GFFNDRRLSE EPNNISFASN AISCYLQAAG LYKNSKIREL LCRILWLISI DDASGMLTNA3120
3121 FDSFRGEIPV WYWITFIPQL LTSLSHKEAN MVRHILIRIA KSYPQALHFQ LRTTKEDFAV3180
3181 IQRQTMAVMG DKPDTNDRNG RRQPWEYLQE LNNILKTAYP LLALSLESLV AQINDRFKST3240
3241 TDEDLFRLIN VLLIDGTLNY NRLPFPRKNP KLPENTEKNL VKFSTTLLAP YIRPKFNADF3300
3301 IDNKPDYETY IKRLRYWRRR LENKLDRASK KENLEVLCPH LSNFHHQKFE DIEIPGQYLL3360
3361 NKDNNVHFIK IARFLPTVDF VRGTHSSYRR LMIRGHDGSV HSFAVQYPAV RHSRREERMF3420
3421 QLYRLFNKSL SKNVETRRRS IQFNLPIAIP LSPQVRIMND SVSFTTLHEI HNEFCKKKGF3480
3481 DPDDIQDFMA DKLNAAHDDA LPAPDMTILK VEIFNSIQTM FVPSNVLKDH FTSLFTQFED3540
3541 FWLFRKQFAS QYSSFVFMSY MMMINNRTPH KIHVDKTSGN VFTLEMLPSR FPYERVKPLL3600
3601 KNHDLSLPPD SPIFHNNEPV PFRLTPNIQS LIGDSALEGI FAVNLFTISR ALIEPDNELN3660
3661 TYLALFIRDE IISWFSNLHR PIIENPQLRE MVQTNVDLII RKVAQLGHLN STPTVTTQFI3720
3721 LDCIGSAVSP RNLARTDVNF MPWF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
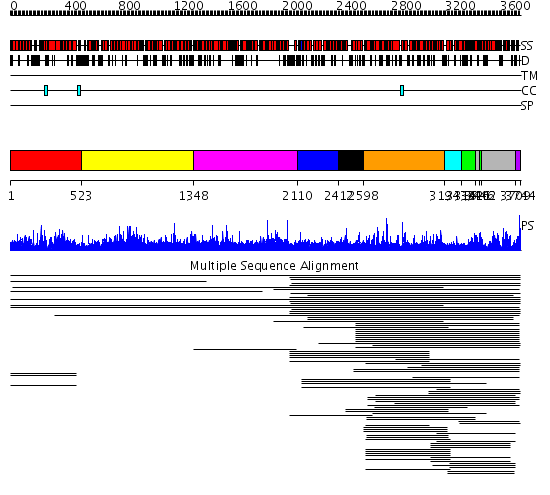
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..522] | 2.009859 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [523..1347] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [1348..2109] | 1.016001 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [2110..2411] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [2412..2597] | 1.031983 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [2598..3193] | 204.744727 | FAT domain No confident structure predictions are available. | |
| 7 | View Details | [3194..3315] | 79.0 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain | |
| 8 | View Details | [3316..3420] [3446..3461] |
79.0 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain | |
| 9 | View Details | [3421..3445] [3462..3708] |
79.0 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain | |
| 10 | View Details | [3709..3744] | 11.79588 | FATC domain No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)