













| General Information: |
|
| Name(s) found: |
ESA1 /
YOR244W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 445 amino acids |
Gene Ontology: |
|
| Cellular Component: |
NuA4 histone acetyltransferase complex
[IPI Piccolo NuA4 histone acetyltransferase complex [IDA histone acetyltransferase complex [IDA |
| Biological Process: |
DNA repair
[IDA RNA elongation [IDA histone acetylation [IDA regulation of transcription from RNA polymerase II promoter [IMP |
| Molecular Function: |
histone acetyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSHDGKEEPG IAKKINSVDD IIIKCQCWVQ KNDEERLAEI LSINTRKAPP KFYVHYVNYN 60
61 KRLDEWITTD RINLDKEVLY PKLKATDEDN KKQKKKKATN TSETPQDSLQ DGVDGFSREN 120
121 TDVMDLDNLN VQGIKDENIS HEDEIKKLRT SGSMTQNPHE VARVRNLNRI IMGKYEIEPW 180
181 YFSPYPIELT DEDFIYIDDF TLQYFGSKKQ YERYRKKCTL RHPPGNEIYR DDYVSFFEID 240
241 GRKQRTWCRN LCLLSKLFLD HKTLYYDVDP FLFYCMTRRD ELGHHLVGYF SKEKESADGY 300
301 NVACILTLPQ YQRMGYGKLL IEFSYELSKK ENKVGSPEKP LSDLGLLSYR AYWSDTLITL 360
361 LVEHQKEITI DEISSMTSMT TTDILHTAKT LNILRYYKGQ HIIFLNEDIL DRYNRLKAKK 420
421 RRTIDPNRLI WKPPVFTASQ LRFAW |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ARP4 EAF1 EAF3 EAF5 EAF6 EAF7 EPL1 ESA1 SWC4 TRA1 YAF9 YNG2 |
| View Details | Hazbun TR, et al. (2003) |

|
ARP4 EAF1 EAF5 EPL1 ESA1 RVB1 RVB2 SWC4 SWR1 TRA1 YAF9 YNG2 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
ESA1 TRA1 |
| View Details | Krogan NJ, et al. (2006) |

|
ARP4 ARP5 ARP6 ARP8 CCA1 EAF1 EAF5 EAF6 EAF7 EPL1 ESA1 IES1 IES2 IES3 IES4 IES5 IES6 INO80 NHP10 NTO1 PIH1 RVB1 RVB2 SAS3 SWC3 SWC4 SWC5 SWC7 SWR1 TAH1 TRA1 VPS71 VPS72 YAF9 YNG2 |
| View Details | Gavin AC, et al. (2006) |

|
EPL1 ESA1 |
| View Details | Gavin AC, et al. (2002) |
|
ACT1 ADH1 ARP4 EAF1 EAF3 EAF5 EPL1 ESA1 HHF1, HHF2 TRA1 YAF9 YAP1 |
| View Details | Ho Y, et al. (2002) |
|
ARP4 ATE1 CDC33 CKA2 EAF1 EPL1 ESA1 HRR25 POR1 SAP185 SIT4 TAF14 TAF4 TIP41 TRA1 TUF1 YRA1 |
| View Details | Qiu et al. (2008) |
|
AHC1 ARP4 EPL1 ESA1 GCN5 TRA1 YNG1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
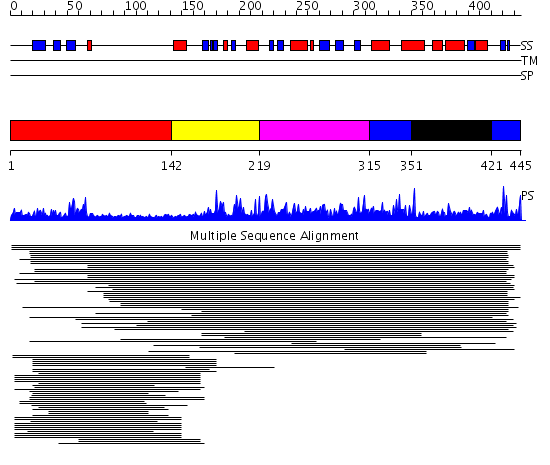
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..141] | 1.5 | Heterochromatin protein 1, HP1 | |
| 2 | View Details | [142..218] | 1714.0 | Histone acetyltransferase ESA1 | |
| 3 | View Details | [219..314] | 1714.0 | Histone acetyltransferase ESA1 | |
| 4 | View Details | [315..350] [421..445] |
1714.0 | Histone acetyltransferase ESA1 | |
| 5 | View Details | [351..420] | 1714.0 | Histone acetyltransferase ESA1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)