













| General Information: |
|
| Name(s) found: |
SWC4 /
YGR002C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 476 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA NuA4 histone acetyltransferase complex [IPI Swr1 complex [IDA histone acetyltransferase complex [IPI |
| Biological Process: |
chromatin modification
[IPI DNA repair [IDA chromatin remodeling [IDA histone exchange [IPI histone acetylation [IPI |
| Molecular Function: |
DNA binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSDIFDVL NIKQKSRSPT NGQVSVPSSS AANRPKPQVT GMQRELFNLL GENQPPVVIK 60
61 SGNNFKEKML STSKPSPWSF VEFKANNSVT LRHWVKGSKE LIGDTPKESP YSKFNQHLSI 120
121 PSFTKEEYEA FMNENEGTQK SVESEKNHNE NFTNEKKDES KNSWSFEEIE YLFNLCKKYD 180
181 LRWFLIFDRY SYNNSRTLED LKEKFYYTCR NYFKASDPSN PLLSSLNFSA EKEIERKKYL 240
241 QRLLSRSAAE IAEEEALVVE SKKFEMAAKR TLAERESLLR LLDSPHSDQT ITQYLTSQGM 300
301 SQLYNALLAD KTRKRKHDLN IPENPWMKQQ QQFAQHRQLQ QLNVKKSEVK ENLSPKKTKR 360
361 QRQEMQTALK RKSESAYAEQ LLKDFNSDER KALGVITHGE KLSPGVYLRS TKLSTFKPAL 420
421 QNKILAILQE LSLPSRPVMP SFDVMERQEE LLKKINTLID LKKHVDKYEA GMSITK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ARP4 ARP6 BDF1 EAF1 EAF3 EAF5 EAF6 EAF7 EPL1 ESA1 RVB1 RVB2 SRP1 SWC3 SWC4 SWC5 SWC7 SWR1 VPS72 YAF9 YNG2 |
| View Details | Hazbun TR, et al. (2003) |

|
ARP4 EAF1 EAF5 EPL1 ESA1 RVB1 RVB2 SWC4 SWR1 TRA1 YAF9 YNG2 |
| View Details | Krogan NJ, et al. (2006) |

|
ARP4 ARP5 ARP6 ARP8 CCA1 EAF1 EAF5 EAF6 EAF7 EPL1 ESA1 IES1 IES2 IES3 IES4 IES5 IES6 INO80 NHP10 NTO1 PIH1 RVB1 RVB2 SAS3 SWC3 SWC4 SWC5 SWC7 SWR1 TAH1 TRA1 VPS71 VPS72 YAF9 YNG2 |
| View Details | Ho Y, et al. (2002) |
|
ARP4 FAA2 HFI1 RPO31 SPT7 STB4 SWC4 SWI5 TRA1 |
| View Details | Qiu et al. (2008) |
|
ARP4 ARP6 EPL1 SPT3 SWC4 TAF10 TAF12 VPS71 VPS72 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Hazbun TR, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
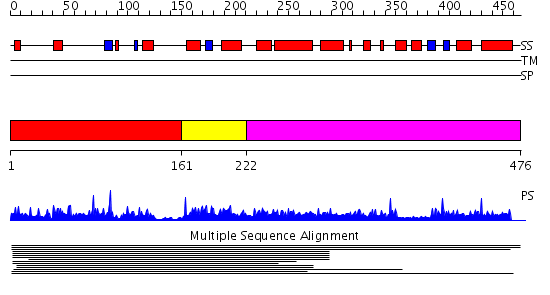
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..160] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [161..221] | 3.568636 | Myb-like DNA-binding domain No confident structure predictions are available. | |
| 3 | View Details | [222..476] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)