













| General Information: |
|
| Name(s) found: |
SWI5 /
YDR146C
[SGD]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 709 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
regulation of transcription involved in G1 phase of mitotic cell cycle
[IDA |
| Molecular Function: |
transcription activator activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTSNSWFDA SKVQSLNFDL QTNSYYSNAR GSDPSSYAIE GEYKTLATDD LGNILNLNYG 60
61 ETNEVIMNEI NDLNLPLGPL SDEKSVKVST FSELIGNDWQ SMNFDLENNS REVTLNATSL 120
121 LNENRLNQDS GMTVYQKTMS DKPHDEKKIS MADNLLSTIN KSEINKGFDR NLGELLLQQQ 180
181 QELREQLRAQ QEANKKLELE LKQTQYKQQQ LQATLENSDG PQFLSPKRKI SPASENVEDV 240
241 YANSLSPMIS PPMSNTSFTG SPSRRNNRQK YCLQRKNSSG TVGPLCFQEL NEGFNDSLIS 300
301 PKKIRSNPNE NLSSKTKFIT PFTPKSRVSS ATSNSANITP NNLRLDFKIN VEDQESEYSE 360
361 KPLGLGIELL GKPGPSPTKS VSLKSASVDI MPTIPGSVNN TPSVNKVSLS SSYIDQYTPR 420
421 GKQLHFSSIS ENALGINAAT PHLKPPSQQA RHREGVFNDL DPNVLTKNTD NEGDDNEENE 480
481 PESRFVISET PSPVLKSQSK YEGRSPQFGT HIKEINTYTT NSPSKITRKL TTLPRGSIDK 540
541 YVKEMPDKTF ECLFPGCTKT FKRRYNIRSH IQTHLEDRPY SCDHPGCDKA FVRNHDLIRH 600
601 KKSHQEKAYA CPCGKKFNRE DALVVHRSRM ICSGGKKYEN VVIKRSPRKR GRPRKDGTSS 660
661 VSSSPIKENI NKDHNGQLMF KLEDQLRRER SYDGNGTGIM VSPMKTNQR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
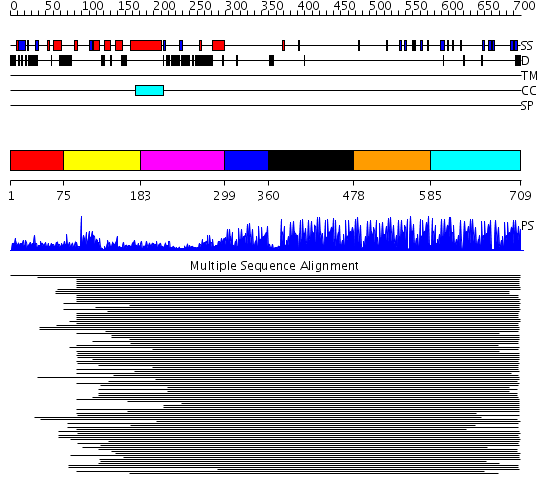
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..74] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [75..182] | 42.524997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [183..298] | 6.848995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [299..359] | 3.69897 | Enhancer binding protein | |
| 5 | View Details | [360..477] | 2.961995 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [478..584] | 126.228787 | Five-finger GLI1 | |
| 7 | View Details | [585..709] | 117.39794 | Designed zinc finger protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 |
|
|||||||||||||||
| 4 | No functions predicted. | |||||||||||||||
| 5 | No functions predicted. | |||||||||||||||
| 6 | No functions predicted. | |||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)