













| General Information: |
|
| Name(s) found: |
RPA135 /
YPR010C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1203 amino acids |
Gene Ontology: |
|
| Cellular Component: |
DNA-directed RNA polymerase I complex
[TAS |
| Biological Process: |
transcription from RNA polymerase I promoter
[TAS]
ribosome biogenesis [RCA |
| Molecular Function: |
DNA-directed RNA polymerase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKVIKPPGQ ARTADFRTLE RESRFINPPK DKSAFPLLQE AVQPHIGSFN ALTEGPDGGL 60
61 LNLGVKDIGE KVIFDGKPLN SEDEISNSGY LGNKLSVSVE QVSIAKPMSN DGVSSAVERK 120
121 VYPSESRQRL TSYRGKLLLK LKWSVNNGEE NLFEVRDCGG LPVMLQSNRC HLNKMSPYEL 180
181 VQHKEESDEI GGYFIVNGIE KLIRMLIVQR RNHPMAIIRP SFANRGASYS HYGIQIRSVR 240
241 PDQTSQTNVL HYLNDGQVTF RFSWRKNEYL VPVVMILKAL CHTSDREIFD GIIGNDVKDS 300
301 FLTDRLELLL RGFKKRYPHL QNRTQVLQYL GDKFRVVFQA SPDQSDLEVG QEVLDRIVLV 360
361 HLGKDGSQDK FRMLLFMIRK LYSLVAGECS PDNPDATQHQ EVLLGGFLYG MILKEKIDEY 420
421 LQNIIAQVRM DINRGMAINF KDKRYMSRVL MRVNENIGSK MQYFLSTGNL VSQSGLDLQQ 480
481 VSGYTVVAEK INFYRFISHF RMVHRGSFFA QLKTTTVRKL LPESWGFLCP VHTPDGSPCG 540
541 LLNHFAHKCR ISTQQSDVSR IPSILYSLGV APASHTFAAG PSLCCVQIDG KIIGWVSHEQ 600
601 GKIIADTLRY WKVEGKTPGL PIDLEIGYVP PSTRGQYPGL YLFGGHSRML RPVRYLPLDK 660
661 EDIVGPFEQV YMNIAVTPQE IQNNVHTHVE FTPTNILSIL ANLTPFSDFN QSPRNMYQCQ 720
721 MGKQTMGTPG VALCHRSDNK LYRLQTGQTP IVKANLYDDY GMDNFPNGFN AVVAVISYTG 780
781 YDMDDAMIIN KSADERGFGY GTMYKTEKVD LALNRNRGDP ITQHFGFGND EWPKEWLEKL 840
841 DEDGLPYIGT YVEEGDPICA YFDDTLNKTK IKTYHSSEPA YIEEVNLIGD ESNKFQELQT 900
901 VSIKYRIRRT PQIGDKFSSR HGQKGVCSRK WPTIDMPFSE TGIQPDIIIN PHAFPSRMTI 960
961 GMFVESLAGK AGALHGIAQD STPWIFNEDD TPADYFGEQL AKAGYNYHGN EPMYSGATGE1020
1021 ELRADIYVGV VYYQRLRHMV NDKFQVRSTG PVNSLTMQPV KGRKRHGGIR VGEMERDALI1080
1081 GHGTSFLLQD RLLNSSDYTQ ASVCRECGSI LTTQQSVPRI GSISTVCCRR CSMRFEDAKK1140
1141 LLTKSEDGEK IFIDDSQIWE DGQGNKFVGG NETTTVAIPF VLKYLDSELS AMGIRLRYNV1200
1201 EPK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
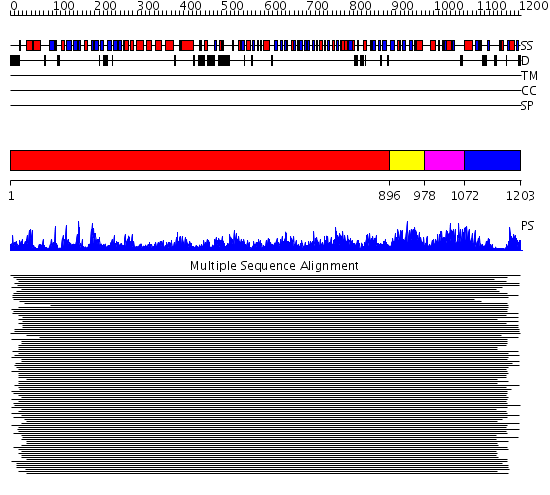
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..895] | 34.361906 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [896..977] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [978..1071] | 3.231973 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [1072..1203] | 28.228997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)