













| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLKNSGSKHS NSKESHSNSS SGIFQNLKRL ANSNATNSNT GSPTYASQQQ HSPVGNEVST 60
61 SPASSSSFRK LNAPSRSTST EARPLNKKST LNTQNLSQYM NGKLSGDVPV SSQHARSHSM 120
121 QSKYSYSKRN SSQASNKLTR QHTGQSHSAS SLLSQGSLTN LSKFTTPDGK IYLEMPSDPY 180
181 EVEVLFEDIM YKRNIFQSLS EDKQEALMGY SIEKKWLIVK QDLQNELKKM RANTTSSSTA 240
241 SRTSMASDHH PILTANSSLS SPKSVLMTSA SSPTSTVYSN SLNHSTTLSS VGTSTSKGKK 300
301 LVSGSLKKQP SLNNIYRGGA ENNTSASTLP GDRTNRPPIH YVQRILADKL TSDEMKDLWV 360
361 TLRTEQLDWV DAFIDHQGHI AMANVLMNSI YKTAPRENLT KELLEKENSF FKCFRVLSML 420
421 SQGLYEFSTH RLMTDTVAEG LFSTKLATRK MATEIFVCML EKKNKSRFEA VLTSLDKKFR 480
481 IGQNLHMIQN FKKMPQYFSH LTLESHLKII QAWLFAVEQT LDGRGKMGSL VGASDEFKNG 540
541 GGENAILEYC QWTMVFINHL CSCSDNINQR MLLRTKLENC GILRIMNKIK LLDYDKVIDQ 600
601 IELYDNNKLD DFNVKLEANN KAFNVDLHDP LSLLKNLWDI CKGTENEKLL VSLVQHLFLS 660
661 SSKLIEENQN SSKLTKQLKL MDSLVTNVSV ASTSDEETNM NMAIQRLYDA MQTDEVARRA 720
721 ILESRALTKK LEEIQAERDS LSEKLSKAEH GLVGQLEDEL HERDRILAKN QRVMQQLEAE 780
781 LEELKKKHLL EKHQQEVELR KMLTILNSRP EESFNKNEGT RGMNSSLNSS EKANIQKVLQ 840
841 DGLSRAKKDY KDDSKKFGMT LQPNKRLKML RMQMENIENE ARQLEMTNFA EFEKDRLEPP 900
901 IHIKKPKVKK MKNKDRKPLV KPQEADVNKL NDLRRALTEI QMESNDISKF NVEERVNELF 960
961 NEKKSLALKR LKELETKYKG FGIDFNVDEI MDSPKKNTGD VETEEDANYA SLDPKTYQKK1020
1021 LDEINRITDQ LLDIQTQTEH EIQVEEDGES DLSSSSSDDE SEEIYQDASP TQELRSEHSE1080
1081 LSSGSGPGSF LDALSQKYGT GQNVTASAAF GENNNGSGIG PLHSKVEKTF MNRLRKSTVS1140
1141 SAPYLEELTQ KVNKVEPYEQ NEDEGLDKKS LPENSTASAA SAFDKAEKDM RQHVENGKQG1200
1201 RVVNHEEDKT ADFSAVSKLN NTDGAEDLST QSSVLSSQPP PPPPPPPPVP AKLFGESLEK1260
1261 EKKSEDDTVK QETTGDSPAP PPPPPPPPPP PMALFGKPKG ETPPPPPLPS VLSSSTDGVI1320
1321 PPAPPMMPAS QIKSAVTSPL LPQSPSLFEK YPRPHKKLKQ LHWEKLDCTD NSIWGTGKAE1380
1381 KFADDLYEKG VLADLEKAFA AREIKSLASK RKEDLQKITF LSRDISQQFG INLHMYSSLS1440
1441 VADLVKKILN CDRDFLQTPS VVEFLSKSEI IEVSVNLARN YAPYSTDWEG VRNLEDAKPP1500
1501 EKDPNDLQRA DQIYLQLMVN LESYWGSRMR ALTVVTSYER EYNELLAKLR KVDKAVSALQ1560
1561 ESDNLRNVFN VILAVGNFMN DTSKQAQGFK LSTLQRLTFI KDTTNSMTFL NYVEKIVRLN1620
1621 YPSFNDFLSE LEPVLDVVKV SIEQLVNDCK DFSQSIVNVE RSVEIGNLSD SSKFHPLDKV1680
1681 LIKTLPVLPE ARKKGDLLED EVKLTIMEFE SLMHTYGEDS GDKFAKISFF KKFADFINEY1740
1741 KKAQAQNLAA EEEERLYIKH KKIVEEQQKR AQEKEKQKEN SNSPSSEGNE EDEAEDRRAV1800
1801 MDKLLEQLKN AGPAKSDPSS ARKRALVRKK YLSEKDNAPQ LLNDLDTEEG SILYSPEAMD1860
1861 PTADTVIHAE SPTPLATRGV MNTSEDLPSP SKTSALEDQE EISDRARMLL KELRGSDTPV1920
1921 KQNSILDEHL EKLRARKERS IGEASTGNRL SFK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2002) |

|
BNI1 CDC48 DOA1 FAL1 MRPS28 NPL4 PDC1 RAI1 REG1 RPA135 RPA190 RPO31 SHP1 UFD1 YDR049W |
| View Details | Ho Y, et al. (2002) |
|
ADR1 ARC15 ARC18 ARC19 ARC35 ARC40 ARP2 ARP3 BNI1 DUR1,2 IMH1 KAP122 MCM4 MCM7 MET18 RPT4 TRP3 YJR029W |
| View Details | Qiu et al. (2008) |
|
ABP1 ABP140 ACT1 AIP1 ARC40 ARP2 BBC1 BEM1 BIK1 BNI1 BNR1 BOI1 BOI2 BUD6 CAP1 CAP2 CDC24 COF1 CRN1 HOF1 HSK3 IQG1 MYO2 MYO4 PAN1 PEA2 PFY1 RVS167 SCP1 SHE3 SLA1 SLA2 SPA2 SPH1 SRV2 TPM2 TWF1 VRP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #22 Asynchronous Prep2-IMAC Phosphopeptide enrichment B1-2 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
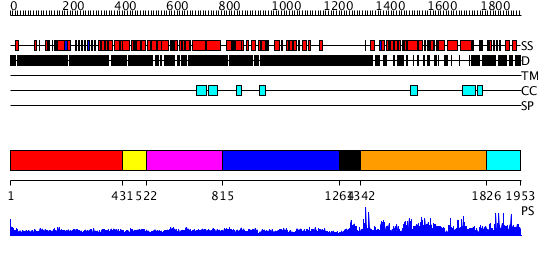
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..430] | 42.09691 | Heavy meromyosin subfragment | |
| 2 | View Details | [431..521] | 10.69897 | Myosin S1 fragment, N-terminal domain; Myosin S1, motor domain | |
| 3 | View Details | [522..814] | 8.0 | Tropomyosin | |
| 4 | View Details | [815..1263] | 1.02 | Structure of Unphosphorylated STAT1 | |
| 5 | View Details | [1264..1341] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1342..1825] | 94.30103 | Bni1p Formin Homology 2 Domain complexed with ATP-actin | |
| 7 | View Details | [1826..1953] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)