













| General Information: |
|
| Name(s) found: |
HOF1 /
YMR032W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 669 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular bud neck contractile ring
[IDA |
| Biological Process: |
cytokinesis
[IMP response to DNA damage stimulus [IMP |
| Molecular Function: |
cytoskeletal protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSYSYEACFW DPNDNGVNIL LGHISQGIRS CDSMILFFKQ RSELEKDYAR RLGAITGKLD 60
61 KDIGTNMDYG KLNETFNVVL SVEKARAQSH SKQSEILFRQ IYTDTKAFAA NLQARYTTLS 120
121 GKIERLRMDK FNKKKGCEVL QKKLQDAQIR FRDLQLNENN MIGAKRVEHN KRELLKWESN 180
181 SQEYKVQLDV LKQEYKASQK FWIHEWAQLS CELQEMENAR ISFLQSKLQQ FATSSMETYI 240
241 LEQTKMDMLT NHLNSFTAAD EISTFSKENG TGRLKHKTSK GDMNSSANWA QMSSISTTSK 300
301 KTESYMDNIR KLSSQLKETE NKRKLASIDK YEKPLPSPEV TMATQFRNST PVIRNETKVV 360
361 ANPTLSLRSS PVQLQSNVDD SVLRQKPDKP RPIVGEEQLK PDEDSKNPDE KGLMVHKRNQ 420
421 SLSSPSESSS SNPTDFSHIK KRQSMESMTT SVSSMANSID DSQRFAKSWN SSNRKRKSMS 480
481 HLQVPSSASS RSDDGGRTPN SAHNLNEDDY NTRRDTSTST ILFKPPVAVR GTSRGHTHRQ 540
541 SMIMQDSSNP IEDALYEMER IQSSSKPGTK TGNIMDERGV VRDRGITVTL PIVTSEGFPV 600
601 IEYAKAMYPL IGNEAPGLAN FHKGDYLLIT EIVNKDWYKG EVYDNDRIDR NHRIGLIPYN 660
661 FIQLLHQGL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
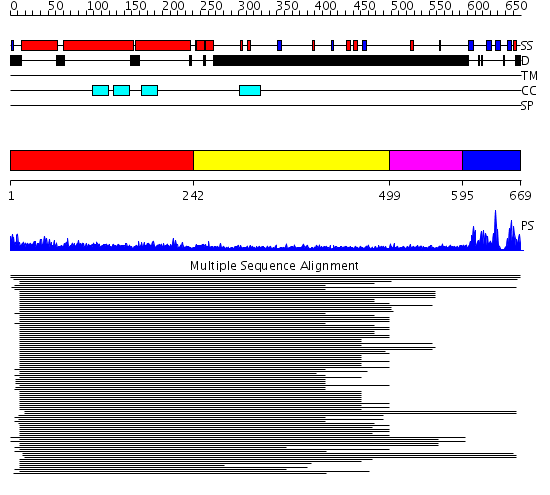
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..241] | 20.0 | Heavy meromyosin subfragment | |
| 2 | View Details | [242..498] | 3.574991 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [499..594] | 1.438996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [595..669] | 18.154902 | SH3 domain from nebulin |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)