













| General Information: |
|
| Name(s) found: |
BEM1 /
YBR200W
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 551 amino acids |
Gene Ontology: |
|
| Cellular Component: |
incipient cellular bud site
[IDA cellular bud tip [IDA cellular bud neck [IDA mating projection tip [TAS |
| Biological Process: |
cell morphogenesis involved in conjugation with cellular fusion
[IGI |
| Molecular Function: |
protein binding
[IDA phosphatidylinositol-3-phosphate binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLKNFKLSKR DSNGSKGRIT SADISTPSHD NGSVIKHIKT VPVRYLSSSS TPVKSQRDSS 60
61 PKNRHNSKDI TSPEKVIKAK YSYQAQTSKE LSFMEGEFFY VSGDEKDWYK ASNPSTGKEG 120
121 VVPKTYFEVF DRTKPSSVNG SNSSSRKVTN DSLNMGSLYA IVLYDFKAEK ADELTTYVGE 180
181 NLFICAHHNC EWFIAKPIGR LGGPGLVPVG FVSIIDIATG YATGNDVIED IKSVNLPTVQ 240
241 EWKSNIARYK ASNISLGSVE QQQQQSITKP QNKSAKLVDG ELLVKASVES FGLEDEKYWF 300
301 LVCCELSNGK TRQLKRYYQD FYDLQVQLLD AFPAEAGKLR DAGGQWSKRI MPYIPGPVPY 360
361 VTNSITKKRK EDLNIYVADL VNLPDYISRS EMVHSLFVVL NNGFDREFER DENQNNIKTL 420
421 QENDTATFAT ASQTSNFAST NQDNTLTGED LKLNKKLSDL SLSGSKQAPA QSTSGLKTTK 480
481 IKFYYKDDIF ALMLKGDTTY KELRSKIAPR IDTDNFKLQT KLFDGSGEEI KTDSQVSNII 540
541 QAKLKISVHD I |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
ABP1 AIP1 ARC40 ARP2 BEM1 BUD6 CAP1 CAP2 COF1 CRN1 DLD2 GLK1 LAS17 OYE2 PAN1 PFY1 RVS167 SAC6 SLA1 SLA2 SRV2 TPM1 TPM2 TWF1 VRP1 |
| View Details | Gavin AC, et al. (2002) |
|
BEM1 BOI2 CDC24 RSC2 |
| View Details | Qiu et al. (2008) |

|
ABP1 ACT1 BEM1 BNI1 BNR1 BOI1 BOI2 BUD6 CDC11 CDC12 CDC24 CDC3 CDC42 COF1 GYL1 HBT1 HOF1 LAS17 LSB3 MLC1 MSB1 MYO1 MYO2 OSH2 PEA2 PFY1 RHO1 ROM2 RVS161 SLA1 SLA2 SPA2 STE20 STE4 TPM1 ZDS1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
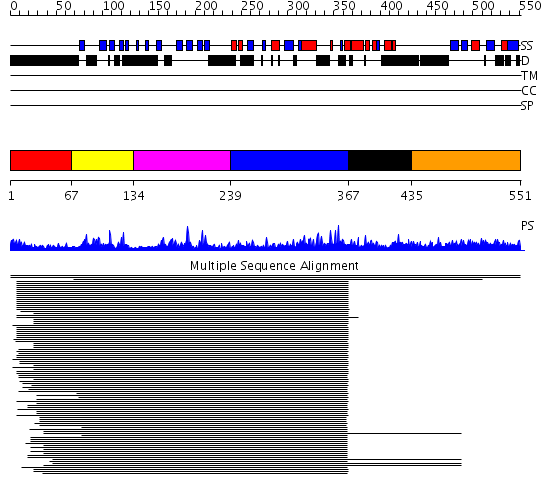
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..66] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [67..133] | 91.24799 | c-src protein tyrosine kinase; c-src tyrosine kinase | |
| 3 | View Details | [134..238] | 91.24799 | c-src protein tyrosine kinase; c-src tyrosine kinase | |
| 4 | View Details | [239..366] | 91.24799 | c-src protein tyrosine kinase; c-src tyrosine kinase | |
| 5 | View Details | [367..434] | 77.156545 | p47phox NADPH oxidase | |
| 6 | View Details | [435..551] | 405.144602 | Bud emergence mediator Bemp1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)