













| General Information: |
|
| Name(s) found: |
FAR1 /
YJL157C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 830 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA membrane [IDA mating projection tip [IDA |
| Biological Process: |
cell cycle arrest
[IMP pheromone-dependent signal transduction involved in conjugation with cellular fusion [IGI |
| Molecular Function: |
cyclin-dependent protein kinase inhibitor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKTPTRVSFE KKIHTPPSGD RDAERSPPKK FLRGLSGKVF RKTPEFKKQQ MPTFGYIEES 60
61 QFTPNLGLMM SKRGNIPKPL NLSKPISPPP SLKKTAGSVA SGFSKTGQLS ALQSPVNITS 120
121 SNKYNIKATN LTTSLLRESI SDSTTMCDTL SDINLTVMDE DYRIDGDSYY EEDSPTFMIS 180
181 LERNIKKCNS QFSPKRYIGE KCLICEESIS STFTGEKVVE STCSHTSHYN CYLMLFETLY 240
241 FQGKFPECKI CGEVSKPKDK DIVPEMVSKL LTGAGAHDDG PSSNMQQQWI DLKTARSFTG 300
301 EFPQFTPQEQ LIRTADISCD GFRTPRLSNS NQFEAVSYLD SPFLNSPFVN KMATTDPFDL 360
361 SDDEKLDCDD EIDESAAEVW FSKTGGEHVM VSVKFQEMRT SDDLGVLQDV NHVDHEELEE 420
421 REKEWKKKID QYIETNVDKD SEFGSLILFD KLMYSDDGEQ WVDNNLVILF SKFLVLFDFE 480
481 EMKILGKIPR DQFYQVIKFN EDVLLCSLKS TNIPEIYLRF NENCEKWLLP KWKYCLENSS 540
541 LETLPLSEIV STVKELSHVN IIGALGAPPD VISAQSHDSR LPWKRLHSDT PLKLIVCLNL 600
601 SHADGELYRK RVLKSVHQIL DGLNTDDLLG IVVVGRDGSG VVGPFGTFIG MINKNWDGWT 660
661 TFLDNLEVVN PNVFRDEKQQ YKVTLQTCER LASTSAYVDT DDHIATGYAK QILVLNGSDV 720
721 VDIEHDQKLK KAFDQLSYHW RYEISQRRMT PLNASIKQFL EELHTKRYLD VTLRLPQATF 780
781 EQVYLGDMAA GEQKTRLIMD EHPHSSLIEI EYFDLVKQQR IHQTLEVPNL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Drees BL, et al. (2001) | ||
| View Screen | 2 | Drees BL, et al. (2001) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
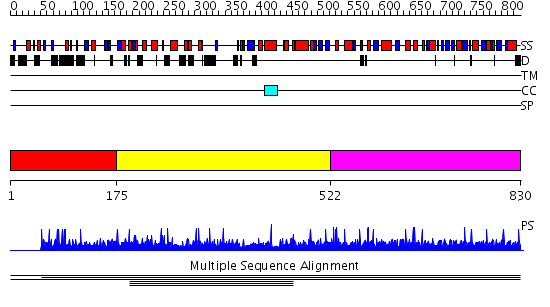
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..174] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [175..521] | 1.002915 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [522..830] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [458..566] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [567..830] | 2.0 | Crystal structure of B. anthracis Protective Antigen complexed with human Anthrax toxin receptor |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)