













| General Information: |
|
| Name(s) found: |
CAR2 /
YLR438W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 424 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
arginine catabolic process
[TAS |
| Molecular Function: |
ornithine-oxo-acid transaminase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEATLSSKQ TIEWENKYSA HNYHPLPVVF HKAKGAHVWD PEGKLYLDFL SAYSAVNQGH 60
61 CHPHIIKALT EQAQTLTLSS RAFHNDVYAQ FAKFVTEFFG FETVLPMNTG AEAVETALKL 120
121 ARRWGYMKKN IPQDKAIILG AEGNFHGRTF GAISLSTDYE DSKLHFGPFV PNVASGHSVH 180
181 KIRYGHAEDF VPILESPEGK NVAAIILEPI QGEAGIVVPP ADYFPKVSAL CRKHNVLLIV 240
241 DEIQTGIGRT GELLCYDHYK AEAKPDIVLL GKALSGGVLP VSCVLSSHDI MSCFTPGSHG 300
301 STFGGNPLAS RVAIAALEVI RDEKLCQRAA QLGSSFIAQL KALQAKSNGI ISEVRGMGLL 360
361 TAIVIDPSKA NGKTAWDLCL LMKDHGLLAK PTHDHIIRLA PPLVISEEDL QTGVETIAKC 420
421 IDLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
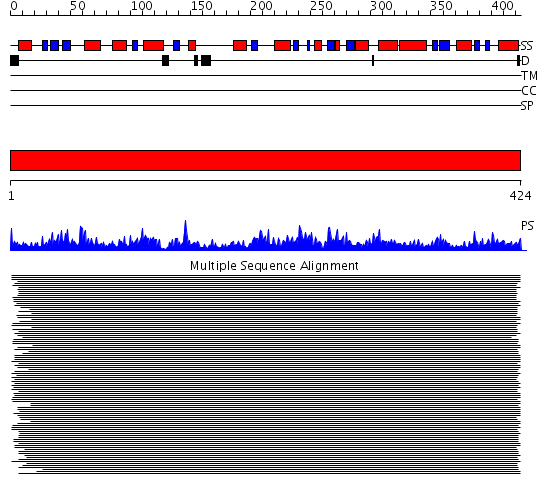
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..424] | 1289.0 | Ornithine aminotransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)