













| General Information: |
|
| Name(s) found: |
SCP160 /
YJL080C
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1222 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum membrane
[IDA polysome [IDA nuclear membrane-endoplasmic reticulum network [IDA |
| Biological Process: |
meiotic telomere clustering
[IMP intracellular mRNA localization [IPI response to drug [IMP chromosome segregation [IMP chromatin silencing at silent mating-type cassette [IMP chromatin silencing at telomere [IGI pheromone-dependent signal transduction involved in conjugation with cellular fusion [IGI |
| Molecular Function: |
RNA binding
[IDA G-protein alpha-subunit binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEEQTAIDS PPSTVEGSVE TVTTIDSPST TASTIAATAE EHPQLEKKPT PLPSLKDLPS 60
61 LGSNAAFANV KVSWGPNMKP AVSNSPSPSP SAPSLTTGLG AKRMRSKNIQ EAFTLDLQSQ 120
121 LSITKPELSR IVQSVKKNHD VSVESTLSKN ARTFLVSGVA ANVHEAKREL VKKLTKPINA 180
181 VIEVPSKCKA SIIGSGGRTI REISDAYEVK INVSKEVNEN SYDEDMDDTT SNVSLFGDFE 240
241 SVNLAKAKIL AIVKEETKNA TIKLVVEDEK YLPYIDVSEF ASDEGDEEVK VQFYKKSGDI 300
301 VILGPREKAK ATKTSIQDYL KKLASNLDEE KVKIPSKFQF LIDAEELKEK YNVIVTFPST 360
361 PDDELVSFVG LRDKVGEAIT YARSSSKSYV VESLDISKAH SKNLTHAKNL IMYFTKYSVL 420
421 KGLEESHPNV KISLPSIQSL PTAETVTIHI SAKSDEANDI KAVRKELISF VNNIPPSETL 480
481 VITDLDYELF GGSIKHCLLA SESSVAFVQF GDYYPNDNSI LLVALTEDED FKPSIEEIQA 540
541 SLNKANESLN SLRTKQNNME TKTYEFSEEV QDSLFKPSSA TWKLIMEDIS EQEGHLQIKL 600
601 HTPEENQLTV RGDEKAAKAA NKIFESILNS PSSKSKMTVN IPANSVARLI GNKGSNLQQI 660
661 REKFACQIDI PNEENNNASK DKTVEVTLTG LEYNLTHAKK YLAAEAKKWA DIITKELIVP 720
721 VKFHGSLIGP HGTYRNRLQE KYNVFINFPR DNEIVTIRGP SRGVNKAHEE LKALLDFEME 780
781 NGHKMVINVP AEHVPRIIGK NGDNINDIRA EYGVEMDFLQ KSTDPKAQET GEVELEITGS 840
841 RQNIKDAAKR VESIVAEASD FVTEVLKIDH KYHKSIVGSG GHILREIISK AGGEEIRNKS 900
901 VDIPNADSEN KDITVQGPQK FVKKVVEEIN KIVKDAENSV TKTIDIPAER KGALIGPGGI 960
961 VRRQLESEFN INLFVPNKDD PSGKITITGA PENVEKAEKK ILNEIIRENF DREVDVPASI1020
1021 YEYVSERGAF IQKLRMDLSV NVRFGNTSKK ANKLARAPIE IPLEKVCGST EGENAEKTKF1080
1081 TIEEVGAPTS SEEGDITMRL TYEPIDLSSI LSDGEEKEVT KDTSNDSAKK EEALDTAVKL1140
1141 IKERIAKAPS ATYAGYVWGA DTRRFNMIVG PGGSNIKKIR EAADVIINVP RKSDKVNDVV1200
1201 YIRGTKAGVE KAGEMVLKSL RR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
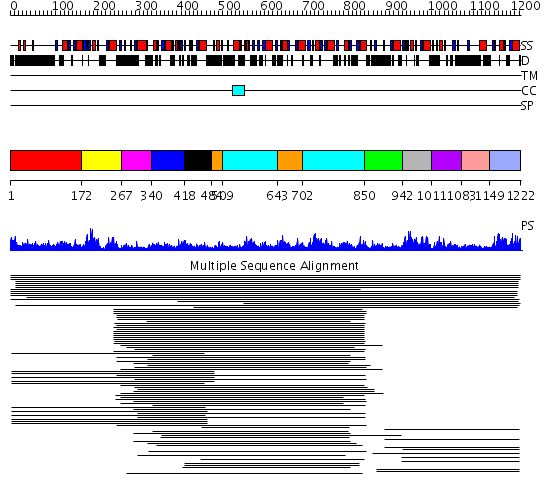
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 1.045986 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [172..266] | 7.522879 | RNA splicing factor 1 | |
| 3 | View Details | [267..339] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [340..417] | 3.221849 | S1 domain of NusA; Transcription factor NusA, C-terminal domains; Transcription factor NusA, N-terminal domain | |
| 5 | View Details | [418..483] | 44.39794 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 6 | View Details | [484..508] [643..701] |
44.39794 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 7 | View Details | [509..642] [702..849] |
44.39794 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 8 | View Details | [850..941] | 11.56 | HnRNP K, KH3 | |
| 9 | View Details | [942..1010] | 32.383689 | Vigilin, KH6 | |
| 10 | View Details | [1011..1082] | 3.39794 | S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase | |
| 11 | View Details | [1083..1148] | 18.045757 | Far upstream binding element, FBP, KH3 and KH4 domains | |
| 12 | View Details | [1149..1222] | 18.045757 | Far upstream binding element, FBP, KH3 and KH4 domains |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)