













| General Information: |
|
| Name(s) found: |
KAP95 /
YLR347C
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 861 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA nuclear pore [IDA |
| Biological Process: |
protein import into nucleus
[TAS nuclear pore complex assembly [IMP |
| Molecular Function: |
protein transmembrane transporter activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTAEFAQLL ENSILSPDQN IRLTSETQLK KLSNDNFLQF AGLSSQVLID ENTKLEGRIL 60
61 AALTLKNELV SKDSVKTQQF AQRWITQVSP EAKNQIKTNA LTALVSIEPR IANAAAQLIA 120
121 AIADIELPHG AWPELMKIMV DNTGAEQPEN VKRASLLALG YMCESADPQS QALVSSSNNI 180
181 LIAIVQGAQS TETSKAVRLA ALNALADSLI FIKNNMEREG ERNYLMQVVC EATQAEDIEV 240
241 QAAAFGCLCK IMSLYYTFMK PYMEQALYAL TIATMKSPND KVASMTVEFW STICEEEIDI 300
301 AYELAQFPQS PLQSYNFALS SIKDVVPNLL NLLTRQNEDP EDDDWNVSMS AGACLQLFAQ 360
361 NCGNHILEPV LEFVEQNITA DNWRNREAAV MAFGSIMDGP DKVQRTYYVH QALPSILNLM 420
421 NDQSLQVKET TAWCIGRIAD SVAESIDPQQ HLPGVVQACL IGLQDHPKVA TNCSWTIINL 480
481 VEQLAEATPS PIYNFYPALV DGLIGAANRI DNEFNARASA FSALTTMVEY ATDTVAETSA 540
541 SISTFVMDKL GQTMSVDENQ LTLEDAQSLQ ELQSNILTVL AAVIRKSPSS VEPVADMLMG 600
601 LFFRLLEKKD SAFIEDDVFY AISALAASLG KGFEKYLETF SPYLLKALNQ VDSPVSITAV 660
661 GFIADISNSL EEDFRRYSDA MMNVLAQMIS NPNARRELKP AVLSVFGDIA SNIGADFIPY 720
721 LNDIMALCVA AQNTKPENGT LEALDYQIKV LEAVLDAYVG IVAGLHDKPE ALFPYVGTIF 780
781 QFIAQVAEDP QLYSEDATSR AAVGLIGDIA AMFPDGSIKQ FYGQDWVIDY IKRTRSGQLF 840
841 SQATKDTARW AREQQKRQLS L |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
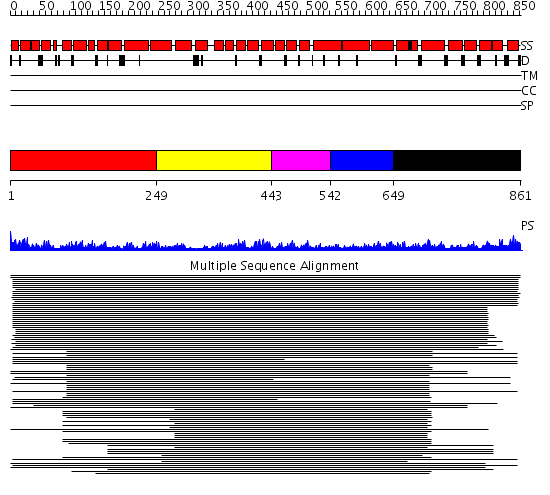
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..248] | 1397.0 | Importin beta | |
| 2 | View Details | [249..442] | 1397.0 | Importin beta | |
| 3 | View Details | [443..541] | 1397.0 | Importin beta | |
| 4 | View Details | [542..648] | 1397.0 | Importin beta | |
| 5 | View Details | [649..861] | 1397.0 | Importin beta |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)