













| General Information: |
|
| Name(s) found: |
TFB4 /
YPR056W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 338 amino acids |
Gene Ontology: |
|
| Cellular Component: |
holo TFIIH complex
[IDA |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter, mitotic
[TAS transcription initiation from RNA polymerase II promoter [TAS |
| Molecular Function: |
general RNA polymerase II transcription factor activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDAISDPTFK HARSRKQVTE ESPSLLTVII EIAPKLWTTF DEEGNEKGSI IKVLEALIVF 60
61 LNAHLAFNSA NKVAVIAAYS QGIKYLYPES TSALKASESE NKTRSDLKII NSDMYRRFRN 120
121 VDETLVEEIY KLFELEKKQI EQNSQRSTLA GAMSAGLTYV NRISKESVTT SLKSRLLVLT 180
181 CGSGSSKDEI FQYIPIMNCI FSATKMKCPI DVVKIGGSKE STFLQQTTDA TNGVYLHVES 240
241 TEGLIQYLAT AMFIDPSLRP IIVKPNHGSV DFRTSCYLTG RVVAVGFICS VCLCVLSIIP 300
301 PGNKCPACDS QFDEHVIAKL KRKPVVPRLK AKKKVTKP |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
SSL1 TFB1 TFB3 TFB4 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
CCL1 KIN28 RAD3 SSL1 SSL2 TFB1 TFB2 TFB3 TFB4 |
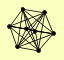
| View Details | Krogan NJ, et al. (2006) |
|
CCL1 ESP1 KIN28 RAD3 SSL1 TFB1 TFB3 TFB4 |
| View Details | Gavin AC, et al. (2006) |

|
SSL1 TFB1 TFB4 |
| View Details | Gavin AC, et al. (2002) |

|
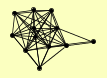
CCL1 CDC48 DCS2 DNA2 ECM29 FAP1 FPR1 KAP95 KIN28 KRS1 MEC1 MPH1 MRPS28 MSH2 MSH6 PGK1 RAD23 RAD34 RAD4 RAD52 RFA1 RFA2 RPN10 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT6 SPT7 SRO7 SSL1 TFA1 TFB1 TFB3 TFB4 TIF6 |
| View Details | Qiu et al. (2008) |
|
CCL1 RAD3 SSL1 SSL2 TAF11 TFA1 TFA2 TFB1 TFB2 TFB3 TFB4 TFB5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
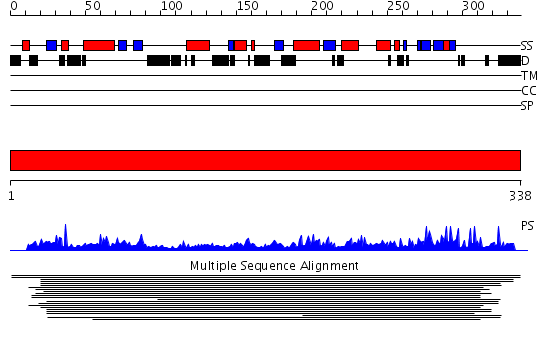
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..338] | 188.552842 | Transcription factor Tfb4 No confident structure predictions are available. | |
| 2 | View Details | [240..338] | 0.954 | No description for 2ozyA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.65 |
Source: Reynolds et al. (2008)