













| General Information: |
|
| Name(s) found: |
RAD3 /
YER171W
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 778 amino acids |
Gene Ontology: |
|
| Cellular Component: |
holo TFIIH complex
[IDA nucleotide-excision repair factor 3 complex [IDA |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter, mitotic
[TAS nucleotide-excision repair, DNA duplex unwinding [TAS nucleotide-excision repair [IDA transcription initiation from RNA polymerase II promoter [TAS |
| Molecular Function: |
DNA helicase activity
[TAS general RNA polymerase II transcription factor activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKFYIDDLPV LFPYPKIYPE QYNYMCDIKK TLDVGGNSIL EMPSGTGKTV SLLSLTIAYQ 60
61 MHYPEHRKII YCSRTMSEIE KALVELENLM DYRTKELGYQ EDFRGLGLTS RKNLCLHPEV 120
121 SKERKGTVVD EKCRRMTNGQ AKRKLEEDPE ANVELCEYHE NLYNIEVEDY LPKGVFSFEK 180
181 LLKYCEEKTL CPYFIVRRMI SLCNIIIYSY HYLLDPKIAE RVSNEVSKDS IVIFDEAHNI 240
241 DNVCIESLSL DLTTDALRRA TRGANALDER ISEVRKVDSQ KLQDEYEKLV QGLHSADILT 300
301 DQEEPFVETP VLPQDLLTEA IPGNIRRAEH FVSFLKRLIE YLKTRMKVLH VISETPKSFL 360
361 QHLKQLTFIE RKPLRFCSER LSLLVRTLEV TEVEDFTALK DIATFATLIS TYEEGFLLII 420
421 EPYEIENAAV PNPIMRFTCL DASIAIKPVF ERFSSVIITS GTISPLDMYP RMLNFKTVLQ 480
481 KSYAMTLAKK SFLPMIITKG SDQVAISSRF EIRNDPSIVR NYGSMLVEFA KITPDGMVVF 540
541 FPSYLYMESI VSMWQTMGIL DEVWKHKLIL VETPDAQETS LALETYRKAC SNGRGAILLS 600
601 VARGKVSEGI DFDHQYGRTV LMIGIPFQYT ESRILKARLE FMRENYRIRE NDFLSFDAMR 660
661 HAAQCLGRVL RGKDDYGVMV LADRRFSRKR SQLPKWIAQG LSDADLNLST DMAISNTKQF 720
721 LRTMAQPTDP KDQEGVSVWS YEDLIKHQNS RKDQGGFIEN ENKEGEQDED EDEDIEMQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
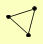
|
MET18 RAD3 TFB3 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
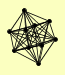
|
CCL1 KIN28 RAD2 RAD3 SSL1 SSL2 TFB1 TFB2 TFB3 TFB4 |
| View Details | Krogan NJ, et al. (2006) |
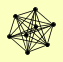
|
CCL1 ESP1 KIN28 RAD3 SSL1 TFB1 TFB3 TFB4 |
| View Details | Gavin AC, et al. (2006) |

|
MET18 RAD3 |
| View Details | Gavin AC, et al. (2002) |
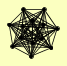
|
ASC1 CLU1 ECM29 GCN1 GCN20 MET18 RAD3 RPN10 RPT3 RPT6 TFP1 TUB3 YDJ1 |
| View Details | Ho Y, et al. (2002) |

|
AAC1 AAC3 ACH1 ACO1 ADH4 ADH6 ATP3 BCY1 BIO3 CCL1 CDC33 ECM10 ERG20 GDI1 HOR2 HXT6 IDH2 KIN28 LSC1 MAM33 MDH1 MET18 PRB1 PTC7 QCR2 RAD26 RAD3 RHR2 RPN1 RPN8 RPT3 SEC53 TEF4 TFB3 TFP1 THI22 TIF1, TIF2 TMA29 YBR184W YSP2 |
| View Details | Qiu et al. (2008) |
|
CCL1 ELC1 KIN28 RAD2 RAD3 SSL1 SSL2 TFB1 TFB2 TFB3 TFB4 TFB5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #21 Asynchronous Prep2-IMAC Phosphopeptide enrichment A1 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
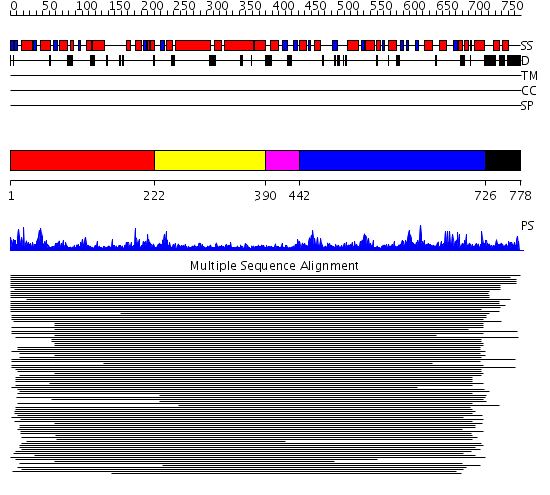
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..221] | 39.0 | Putative DEAD box RNA helicase | |
| 2 | View Details | [222..389] | 39.0 | Putative DEAD box RNA helicase | |
| 3 | View Details | [390..441] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [442..725] | 9.08 | Initiation factor 4a | |
| 5 | View Details | [726..778] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)