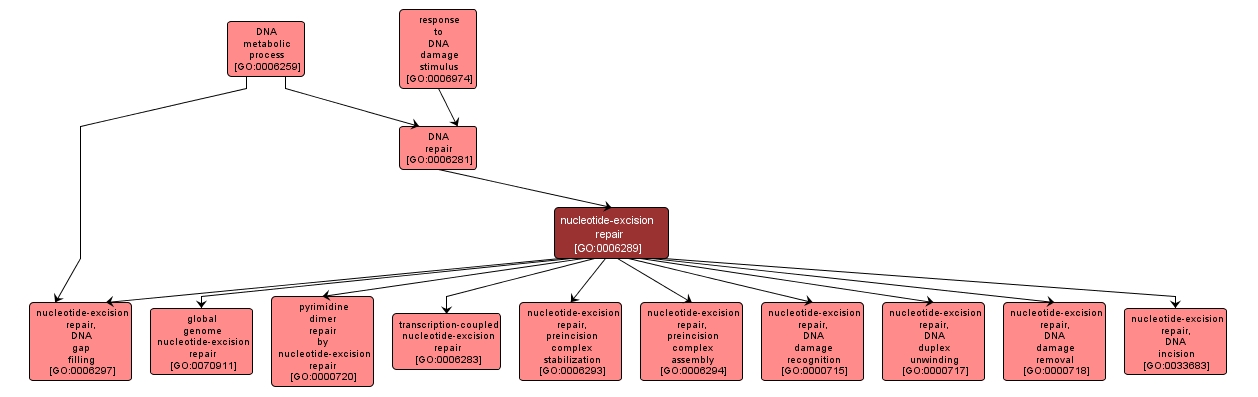
GO TERM SUMMARY
|
| Name: |
nucleotide-excision repair |
| Acc: |
GO:0006289 |
| Aspect: |
Biological Process |
| Desc: |
A DNA repair process in which a small region of the strand surrounding the damage is removed from the DNA helix as an oligonucleotide. The small gap left in the DNA helix is filled in by the sequential action of DNA polymerase and DNA ligase. Nucleotide excision repair recognizes a wide range of substrates, including damage caused by UV irradiation (pyrimidine dimers and 6-4 photoproducts) and chemicals (intrastrand cross-links and bulky adducts). |
Synonyms:
- GO:0045001
- intrastrand cross-link repair
- pyrimidine-dimer repair, DNA damage excision
- NER
- interstrand crosslink repair
|
|

|
INTERACTIVE GO GRAPH
|