













| General Information: |
|
| Name(s) found: |
TEF4 /
YKL081W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 412 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA ribosome [TAS eukaryotic translation elongation factor 1 complex [ISS |
| Biological Process: |
translational elongation
[TAS |
| Molecular Function: |
translation elongation factor activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQGTLYINR SPRNYASEAL ISYFKLDVKI VDLEQSSEFA SLFPLKQAPA FLGPKGLKLT 60
61 EALAIQFYLA NQVADEKERA RLLGSDVIEK SQILRWASLA NSDVMSNIAR PFLSFKGLIP 120
121 YNKKDVDACF VKIDNLAAVF DARLRDYTFV ATENISLGDL HAAGSWAFGL ATILGPEWRA 180
181 KHPHLMRWFN TVAASPIVKT PFAEVKLAEK ALTYTPPKKQ KAEKPKAEKS KAEKKKDEAK 240
241 PADDAAPAKK PKHPLEALGK STFVLDDWKR KYSNDDTRPV ALPWFWEHYN PEEYSIWKVG 300
301 YKYNDELTLT FMSNNLVGGF FNRLSASTKY MFGCLVVYGE NNNNGIVGAV MVRGQDFAPA 360
361 FDVAPDWESY EYTKLDPTKE EDKEFVNNMW AWDKPVVVNG EDKEIVDGKV LK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #35 Asynchronous SPB prep | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
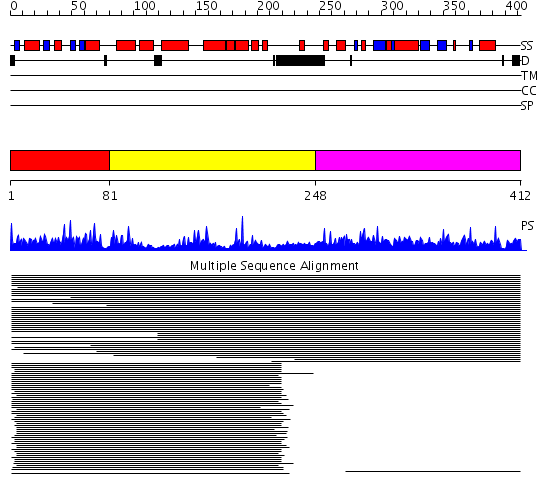
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | 66.533179 | Class phi GST | |
| 2 | View Details | [81..247] | 66.533179 | Class phi GST | |
| 3 | View Details | [248..412] | 81.920819 | Elongation factor 1 gamma, conserved domain No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)