













| General Information: |
|
| Name(s) found: |
PUP1 /
YOR157C
[SGD]
|
| Description(s) found:
Found 37 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 261 amino acids |
Gene Ontology: |
|
| Cellular Component: |
proteasome core complex, beta-subunit complex
[IPI |
| Biological Process: |
ubiquitin-dependent protein catabolic process
[TAS |
| Molecular Function: |
endopeptidase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGLSFDNYQ RNNFLAENSH TQPKATSTGT TIVGVKFNNG VVIAADTRST QGPIVADKNC 60
61 AKLHRISPKI WCAGAGTAAD TEAVTQLIGS NIELHSLYTS REPRVVSALQ MLKQHLFKYQ 120
121 GHIGAYLIVA GVDPTGSHLF SIHAHGSTDV GYYLSLGSGS LAAMAVLESH WKQDLTKEEA 180
181 IKLASDAIQA GIWNDLGSGS NVDVCVMEIG KDAEYLRNYL TPNVREEKQK SYKFPRGTTA 240
241 VLKESIVNIC DIQEEQVDIT A |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
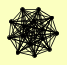
|
BLM10 PRE1 PRE10 PRE2 PRE3 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 SCL1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
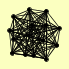
|
PRE1 PRE10 PRE2 PRE3 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 SCL1 UMP1 |
| View Details | Krogan NJ, et al. (2006) |

|
ACK1 CIN8 DCG1 GNA1 MCM6 MLP1 PMD1 PMT4 PRE1 PRE10 PRE2 PRE3 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 RFT1 SCL1 SGA1 SNQ2 TOS8 UMP1 YCR076C YDR179W-A YJL132W YKR070W YLR211C YLR290C |
| View Details | Gavin AC, et al. (2002) |

|
BLM10 CDC55 CIN1 ERG13 GUS1 HHF1, HHF2 HOS2 HOS4 IML1 KAP95 KEL1 LTE1 MYO5 PFK1 PPH21 PPH22 PRE1 PRE10 PRE2 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 RRD2 RTS1 RTS3 SCL1 SET3 SIF2 SNT1 SRP1 TDH2 TDH3 TEF4 TPD3 YBL104C YOR1 YRA1 ZDS1 ZDS2 |
| View Details | Qiu et al. (2008) |

|
PRE1 PRE10 PRE2 PRE3 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 RPN10 RPN12 RPN13 RPT1 RPT2 RPT3 RPT4 RPT6 SCL1 UMP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #21 Asynchronous Prep2-IMAC Phosphopeptide enrichment A1 | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |

Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..261] | 1396.30103 | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST IN COMPLEX WITH THE PROTEASOME ACTIVATOR PA26 FROM TRYPANOSOME BRUCEI AT 3.2 ANGSTROMS RESOLUTION |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.57 |
Source: Reynolds et al. (2008)