













| General Information: |
|
| Name(s) found: |
RTS1 /
YOR014W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 757 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA condensed nuclear chromosome, centromeric region [IDA cytoplasm [IDA cellular bud neck [IDA spindle pole body [IDA protein phosphatase type 2A complex [TAS |
| Biological Process: |
protein amino acid dephosphorylation
[TAS meiotic sister chromatid cohesion, centromeric [IMP translation [ISS |
| Molecular Function: |
protein serine/threonine phosphatase activity
[IPI]
protein phosphatase type 2A regulator activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMRGFKQRLI KKTTGSSSSS SSKKKDKEKE KEKSSTTSST SKKPASASSS SHGTTHSSAS 60
61 STGSKSTTEK GKQSGSVPSQ GKHHSSSTSK TKTATTPSSS SSSSRSSSVS RSGSSSTKKT 120
121 SSRKGQEQSK QSQQPSQSQK QGSSSSSAAI MNPTPVLTVT KDDKSTSGED HAHPTLLGAV 180
181 SAVPSSPISN ASGTAVSSDV ENGNSNNNNM NINTSNTQDA NHASSQSIDI PRSSHSFERL 240
241 PTPTKLNPDT DLELIKTPQR HSSSRFEPSR YTPLTKLPNF NEVSPEERIP LFIAKVDQCN 300
301 TMFDFNDPSF DIQGKEIKRS TLDELIEFLV TNRFTYTNEM YAHVVNMFKI NLFRPIPPPV 360
361 NPVGDIYDPD EDEPVNELAW PHMQAVYEFF LRFVESPDFN HQIAKQYIDQ DFILKLLELF 420
421 DSEDIRERDC LKTTLHRIYG KFLSLRSFIR RSMNNIFLQF IYETEKFNGV AELLEILGSI 480
481 INGFALPLKE EHKVFLVRIL IPLHKVRCLS LYHPQLAYCI VQFLEKDPLL TEEVVMGLLR 540
541 YWPKINSTKE IMFLNEIEDI FEVIEPLEFI KVEVPLFVQL AKCISSPHFQ VAEKVLSYWN 600
601 NEYFLNLCIE NAEVILPIIF PALYELTSQL ELDTANGEDS ISDPYMLVEQ AINSGSWNRA 660
661 IHAMAFKALK IFLETNPVLY ENCNALYLSS VKETQQRKVQ REENWSKLEE YVKNLRINND 720
721 KDQYTIKNPE LRNSFNTASE NNTLNEENEN DCDSEIQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CDC55 PPH21 PPH22 RRD2 RTS1 TPD3 ZDS1 |
| View Details | Krogan NJ, et al. (2006) |

|
AHA1 BUD3 CNS1 CPR6 DIG2 DNM1 DSF1, YNR073C FRT2 GPA2 HOM6 HSC82 HSP104 HSP82 MBB1 MRPL50 PAM1 PPT1 QNS1 RMD8 RTS1 SMT3 STI1 STP4 TDH1 UTP20 YBL104C |
| View Details | Gavin AC, et al. (2002) |

|
BLM10 CDC55 CIN1 ERG13 GUS1 HHF1, HHF2 HOS2 HOS4 IML1 KAP95 KEL1 LTE1 MYO5 PFK1 PPH21 PPH22 PRE1 PRE10 PRE2 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 RRD2 RTS1 RTS3 SCL1 SET3 SIF2 SNT1 SRP1 TDH2 TDH3 TEF4 TPD3 YBL104C YOR1 YRA1 ZDS1 ZDS2 |
| View Details | Ho Y, et al. (2002) |
|
CDC55 GPG1 HEM15 HTA2 MKT1 PPE1 PPH21 PPH22 RPA135 RPB11 RPC40 RTS1 RTS3 RVB1 TAP42 TPD3 |
| View Details | Qiu et al. (2008) |
|
CDC14 CDC55 GIP2 GLC7 PPH21 PPH22 RTS1 RTS3 TPD3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
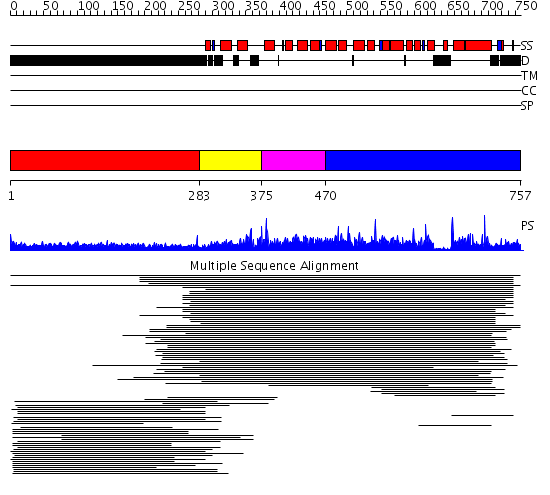
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..282] | 12.27 | Fibrinogen | |
| 2 | View Details | [283..374] | 1.320983 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [375..469] | 8.75 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 4 | View Details | [470..757] | 8.75 | Constant regulatory domain of protein phosphatase 2a, pr65alpha |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)