













| General Information: |
|
| Name(s) found: |
BLM10 /
YFL007W
[SGD]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2143 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA proteasome core complex [IDA proteasome complex [IDA |
| Biological Process: |
protein catabolic process
[IGI proteasome assembly [IMP |
| Molecular Function: |
proteasome activator activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTANNDDDIK SPIPITNKTL SQLKRFERSP GRPSSSQGEI KRKKSRLYAA DGRPHSPLRA 60
61 RSATPTLQDQ KLFNGMDSTS LLNERLQHYT LDYVSDRAQH MKNIYDPSSR WFSRSVRPEF 120
121 PIEEFLPYKT ESHEDQAKYL CHVLVNLYIA ISSLDIQGLI SISSKDLADL KKEVDDLALK 180
181 TDLFRLSNNT AENDLLGNDI ADYDDAEGLE DELDEYFDLA GPDFNATGKI TAKSATIVNV 240
241 NHWTNELKNC LHFDFPVALR KSLATVYYYL SLVQGQKVYR QMHVDMFERL VSLDDDRTNF 300
301 TELLQKQGLL LDHQIMLNFL CEFLPYPDPD YARYELSSKE DLQLFRLLLK HAHNAKPFFD 360
361 KSKESLLVDT MNFLLSSLAP STMMAVMPIV TSVVPYHYHI HSKIIDYFPF CYSIWSSVSA 420
421 NVAIDTHMYD FVGSISKDVH NKILSSEHEK DVVGVEFGEF GIFTDDQMTF MFNRLQGHLR 480
481 TDGQIHSYSR TVKPFVYAIN GSKKDRFFEK LVSLAKAIET FIHPSNNGFW TKPNAKFVHA 540
541 FIKSYHGRVK YEEDICARGV TNGICLTSFC HEEIVEIFLN IISLGSQNKN PDIANYYISC 600
601 FAYLLELDPS NAYLIYDKIL IDLYDTLADQ FINSRHRIIS SLKQFTRVIR FIVMDKLYRV 660
661 HITNVLSMLV SKLDMNDTNL TSNLINGIVS IAAFIPIQDL TGEDDYISFE SDTLPLVQQH 720
721 FYHIKCGESS KTFRVDDELL NNAFKASTTV FQSMLKVYVE KIFQLVDVDL EDSLVTKINQ 780
781 TTMILQESMD DKIFNYFASL LQRNFWSNDS FKEKDPNYEL VTIPLAALVR RNNGLSKELV 840
841 RTLLFHIKEQ IKRGAGSVRS TSEIQQRDVK LVLYLTALND VLRQCHESLL EYSDELITFM 900
901 KYLYDNVTNP PLDVITSIVI HSALATLCTT EITDCRLFPE DSKIPEKDRW GGLQFDPRRF 960
961 DKQHLSFQWH VPSSDEITLS ISILESLSEY CINNVEELMK APRHDSEYGD MIQKYVLVMT1020
1021 HTLSGSSLLF DPDFNKYRTQ SNLSYREKLI LLKNIRENNC DPQELDIDIE QIRSGKDDED1080
1081 YIESKDIEAG LNAGVSDVVQ LRDEFPDELI VDEEVVSEMP SGVNTPIAGT HGTDNSAMSS1140
1141 DLAFRDLDIY TCNYYFGNTT EEKLQNPQYL QVHRVRARIG HFFHKLYVFL STNFENNTNM1200
1201 FQILLHGLKV WFTDLGQETV FNEDPNAFID VDFLENVQSL SHVNEPFTRT NFAIRANSLH1260
1261 QSRVLLHSTN RKASKLENLL LVDIIQLATS LYPDIYKPAQ GTLVHCMKQL VGSYGVVINK1320
1321 IIPSLEKAIK DHDYMKIQVI LNVLLIKKIH RKLMTDYKDI GRLIFLLIEC CRVNELEIGM1380
1381 YADKILTDIV IGIKIPSSVC VISDQAFLPL APPDGTINLQ VEAVKLAKKK KREYYLSLLV1440
1441 DLQDKLLDKL DNEKDMGWKI RMFILRFVTQ IQSNLESKPD KRAVFSIISQ ISTKHPEIIH1500
1501 LVVKSLLSTC NKIISLSDYE YDITRAYKNE FNPSFVEILD TSTTSFPKTF TEEMNNFDNP1560
1561 KYFIDLRAYV GWLCWGRLMY VMSPKALKLN LRENELEVLK TAGHLLTREF LRDVTMNLVQ1620
1621 DNETRGVFSS GNVSFFSLVI LLISSGFCEL NMSDLFELCE SYYNKDDKAS MIMSVEIVAG1680
1681 LVCGSKFMSV SDLDKRDTFI ENFLAKCLDY ELNHDAFEIW STLAWWLPAV VDLRRSKTFF1740
1741 CHFINADGMF DRESDAATHQ TSKIYMLRSI LMSMEFRAPD VGKLFDELVF DHPYDQVRQA1800
1801 VAKLLTTLVQ NQSNPSISDP TTLLEAERND PDGLGLPLKS VPEKVDAYIK KQFEIIKNLE1860
1861 DSVVGLNPQQ FIKTDYFYRT STIFYWIKEM ARGPNKVLLV PYLVDYVLPF LIGLVKHKDV1920
1921 CALASLDPVR LYAGLGYMPI RKNHVAAIVD YVCSSNVALS SNQTKLQLAF IQHFLSAELL1980
1981 QLTEEEKNKI LEFVVSNLYN EQFVEVRVRA ASILSDIVHN WKEEQPLLSL IERFAKGLDV2040
2041 NKYTSKERQK LSKTDIKIHG NVLGLGAIIS AFPYVFPLPP WIPKQLSNLS SWARTSGMTG2100
2101 QAAKNTISEF KKVRADTWKF DRASFNTEEL EDLEGVLWRS YYA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
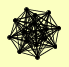
|
BLM10 PRE1 PRE10 PRE2 PRE3 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 SCL1 |
| View Details | Gavin AC, et al. (2002) |

|
BLM10 CDC55 CDC6 CIN1 DAM1 DBP9 ECM29 ERG13 EST3 GFA1 GUS1 HHF1, HHF2 HOS2 HOS4 IML1 INO4 KAP95 KEL1 LTE1 LYS12 MDS3 MLH2 MYO5 NUD1 PDA1 PDB1 PFK1 PGK1 PPH21 PPH22 PRE1 PRE10 PRE2 PRE3 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PSE1 PUP1 PUP2 PUP3 RGR1 RPN10 RPN11 RPN12 RPN13 RPN3 RPN5 RPN6 RPN7 RPN8 RPN9 RPT1 RPT2 RPT3 RPT4 RPT5 RPT6 RRD2 RRP12 RTS1 RTS3 SCL1 SET3 SIF2 SLX9 SNT1 SPA2 SRP1 TDH2 TDH3 TEF4 TPD3 UBP6 ULP1 YBL104C YFL006W YMR310C YOR1 YRA1 ZDS1 ZDS2 |
| View Details | Ho Y, et al. (2002) |
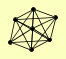
|
BLM10 SEC53 SIR2 SIR3 SIR4 SRP1 YFL006W |
| View Details | Qiu et al. (2008) |

|
BLM10 PRE10 UMP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
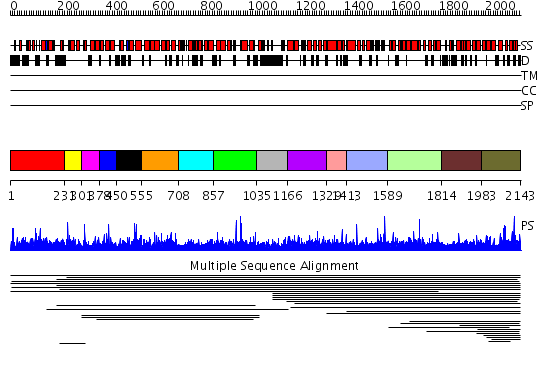
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..230] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [231..300] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [301..377] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [378..449] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [450..554] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [555..707] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [708..856] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [857..1034] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1035..1165] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1166..1328] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1329..1412] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1413..1588] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [1589..1813] | N/A | No confident structure predictions are available. | |
| 14 | View Details | [1814..1982] | N/A | No confident structure predictions are available. | |
| 15 | View Details | [1983..2143] | 3.023948 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 10 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 13 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 14 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 15 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.57 |
Source: Reynolds et al. (2008)