













| General Information: |
|
| Name(s) found: |
SNT1 /
YCR033W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1226 amino acids |
Gene Ontology: |
|
| Cellular Component: |
histone deacetylase complex
[IPI |
| Biological Process: |
histone deacetylation
[IDA negative regulation of meiosis [IPI |
| Molecular Function: |
NAD-dependent histone deacetylase activity
[IPI NAD-independent histone deacetylase activity [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGYPPPTRRL GDKKRYHYSN NPNRRHPSAV YSKNSFPKSS NNGFVSSPTA DNSTNPSVTP 60
61 STASVPLPTA APGSTFGIEA PRPSRYDPSS VSRPSSSSYS STRKIGSRYN PDVERSSSTT 120
121 SSTPESMNTS TITHTNTDIG NSRYSRKTMS RYNPQSTSST NVTHFPSALS NAPPFYVANG 180
181 SSRRPRSMDD YSPDVTNKLE TNNVSSVNNN SPHSYYSRSN KWRSIGTPSR PPFDNHVGNM 240
241 TTTSNTNSIH QREPFWKANS TTILKSTHSQ SSPSLHTKKF HDANKLDKPE ASVKVETPSK 300
301 DETKAISYHD NNFPPRKSVS KPNAPLEPDN IKVGEEDALG KKEVHKSGRE IAKEHPTPVK 360
361 MKEHDELEAR AKKVSKINID GKQDEIWTTA KTVASAVEVS KESQKELTRS VERKESPEIR 420
421 DYERAYDPKA LKTDVTKLTV DNDNKSYEEP LEKVEGCIFP LPKAETRLWE LKNQKRNKII 480
481 SEQKYLLKKA IRNFSEYPFY AQNKLIHQQA TGLILTKIIS KIKKEEHLKK INLKHDYFDL 540
541 QKKYEKECEI LTKLSENLRK EEIENKRKEH ELMEQKRREE GIETEKEKSL RHPSSSSSSR 600
601 RRNRADFVDD AEMENVLLQI DPNYKHYQAA ATIPPLILDP IRKHSYKFCD VNNLVTDKKL 660
661 WASRILKDAS DNFTDHEHSL FLEGYLIHPK KFGKISHYMG GLRSPEECVL HYYRTKKTVN 720
721 YKQLLIDKNK KRKMSAAAKR RKRKERSNDE EVEVDESKEE STNTIEKEEK SENNAEENVQ 780
781 PVLVQGSEVK GDPLGTPEKV ENMIEQRGEE FAGELENAER VNDLKRAHDE VGEESNKSSV 840
841 IETNNGVQIM DPKGAVQNGY YPEETKELDF SLENALQRKK HKSAPEHKTS YWSVRESQLF 900
901 PELLKEFGSQ WSLISEKLGT KSTTMVRNYY QRNAARNGWK LLVDETDLKR DGTSSESVQQ 960
961 SQILIQPERP NINAYSNIPP QQRPALGYFV GQPTHGHNTS ISSIDGSIRP FGPDFHRDTF1020
1021 SKISAPLTTL PPPRLPSIQF PRSEMAEPTV TDLRNRPLDH IDTLADAASS VTNNQNFSNE1080
1081 RNAIDIGRKS TTISNLLNNS DRSMKSSFQS ASRHEAQLED TPSMNNIVVQ EIKPNITTPR1140
1141 SSSISALLNP VNGNGQSNPD GRPLLPFQHA ISQGTPTFPL PAPRTSPISR APPKFNFSND1200
1201 PLAALAAVAS APDAMSSFLS KKENNN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CPR1 HOS2 HOS4 SET3 SIF2 SNT1 |
| View Details | Krogan NJ, et al. (2006) |
|
AKL1 HOS4 HST1 RFM1 SET3 SIF2 SNT1 SUM1 |
| View Details | Gavin AC, et al. (2006) |
|
CPR1 HOS2 HOS4 SET3 SIF2 SNT1 |
| View Details | Gavin AC, et al. (2002) |

|
BLM10 CDC55 CIN1 CPR1 ERG13 GUS1 HHF1, HHF2 HOS2 HOS4 IML1 KAP95 KEL1 LTE1 MYO5 PFK1 PPH21 PPH22 PRE1 PRE10 PRE2 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 RRD2 RTS1 RTS3 RVB2 SCL1 SET3 SIF2 SNF2 SNT1 SRP1 TDH2 TDH3 TEF4 TPD3 YBL104C YOR1 YRA1 ZDS1 ZDS2 |
| View Details | Ho Y, et al. (2002) |
|
AVT2 HOS4 SIF2 SNT1 TFP1 TRM3 UTP21 YMR155W YRF1-3, YRF1-7 ZDS2 |
| View Details | Qiu et al. (2008) |
|
HDA1 HDA2 HDA3 HOS1 HOS2 HOS4 HST1 RPD3 SDS3 SET3 SIF2 SIN3 SNT1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
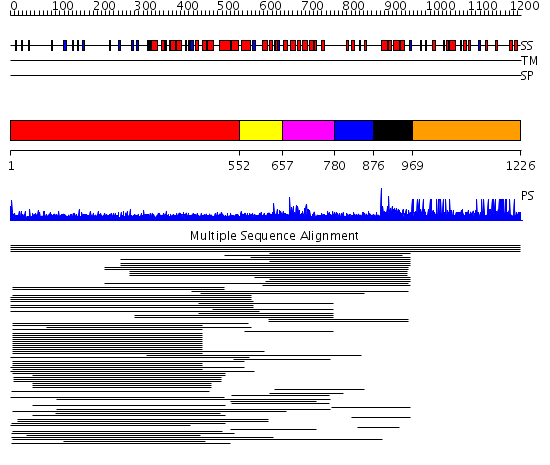
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..551] | 40.291998 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [552..656] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [657..779] | 11.119186 | Myb-like DNA-binding domain No confident structure predictions are available. | |
| 4 | View Details | [780..875] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [876..968] | 10.49485 | Myb-like DNA-binding domain No confident structure predictions are available. | |
| 6 | View Details | [969..1226] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [831..965] | 38.69897 | No description for 2iw5B was found. | |
| 8 | View Details | [966..1226] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)