













| General Information: |
|
| Name(s) found: |
HDA2 /
YDR295C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 674 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA histone deacetylase complex [IMP |
| Biological Process: |
telomere maintenance
[IMP regulation of transcription, DNA-dependent [IDA histone deacetylation [IDA |
| Molecular Function: |
histone deacetylase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRKNSKKLK VYYLPVTLTQ FQKDLSEILI SLHAKSFKAS IIGEPQADAV NKPSGLPAGP 60
61 ETHPYPTLSQ RQLTYIFDSN IRAIANHPSL LVDHYMPRQL LRMEPTESSI AGSHKFQVLN 120
121 QLINSICFRD REGSPNEVIK CAIIAHSIKE LDLLEGLILG KKFRTKRLSG TSLYNEKHKF 180
181 PNLPTVDSTI NKDGTPNSVS STSSNSNSTS YTGYSKDDYD YSVKRNLKKR KINTDDWLFL 240
241 ATTKHLKHDQ YLLANYDIDM IISFDPMLEV ELPALQVLRN NANKDIPIIK LLVQNSPDHY 300
301 LLDSEIKNSS VKSSHLSNNG HVDDSQEYEE IKSSLLYFLQ ARNAPVNNCE IDYIKLVKCC 360
361 LEGKDCNNIL PVLDLITLDE ASKDSSDSGF WQPQLTKLQY SSTELPLWDG PLDIKTYQTE 420
421 LMHRAVIRLR DIQDEYAKGT VPLYEKRLNE TQRQNQLDEI KNSVGLTFKK KQEVEKSIND 480
481 SEKRLKHAMT ESTKLQNKIN HLLKNRQELE NFNKLPSNTI SSENHLEEGS ALADKLKEYI 540
541 DKNATLFNKL KELQQANAEK SKLNDELRSK YQIESSKAAE SAQTLKILQE SMKSLENEVN 600
601 GPLTKFSTES LKKELERLQN DFQSLKARNK FLKNYITLMN RQYDLKNKNN VQVEKAAANG 660
661 TRFRSTRSNT PNYT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
HDA1 HDA2 HDA3 |
| View Details | Krogan NJ, et al. (2006) |

|
HDA1 HDA2 HDA3 |
| View Details | Gavin AC, et al. (2006) |

|
HDA1 HDA2 HDA3 PRO2 |
| View Details | Gavin AC, et al. (2002) |

|
CMR1 DNA2 HDA1 HDA2 HDA3 HSC82 KAP95 LCD1 MEC1 MGM101 MLH1 MPH1 MSH2 MSH3 MSH6 OLA1 PGK1 PMS1 PRO2 RAD16 RAD52 RFA1 RFA2 RFA3 RIM1 RNH1 RSC8 RVB2 SGS1 SHS1 TIF1, TIF2 TOP3 VAS1 VPS1 YHR122W |
| View Details | Qiu et al. (2008) |

|
HDA1 HDA2 HDA3 HOS1 HOS4 RPD3 SDS3 SET3 SIN3 SNT1 STB2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
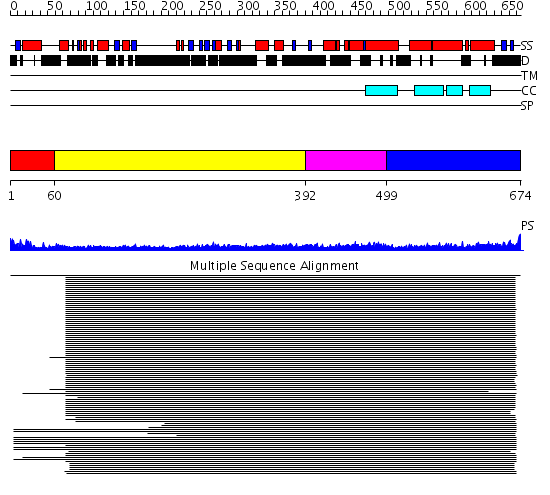
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..59] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [60..391] | 17.154902 | Heavy meromyosin subfragment | |
| 3 | View Details | [392..498] | 8.69897 | Tropomyosin | |
| 4 | View Details | [499..674] | 4.212997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)