













| General Information: |
|
| Name(s) found: |
MSH3 /
YCR092C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1047 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
removal of nonhomologous ends
[IGI DNA recombination [IMP meiotic mismatch repair [IMP mismatch repair [IMP mitotic recombination [IMP |
| Molecular Function: |
double-strand/single-strand DNA junction binding
[IDA DNA insertion or deletion binding [IDA loop DNA binding [IDA Y-form DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVIGNEPKLV LLRAKSSANR FILLNLLTIM AGQPTISRFF KKAVKSELTH KQEQEVAVGN 60
61 GAGSESICLD TDEEDNLSSV ASTTVTNDSF PLKGSVSSKN SKNSEKTSGT STTFNDIDFA 120
121 KKLDRIMKRR SDENVEAEDD EEEGEEDFVK KKARKSPTAK LTPLDKQVKD LKMHHRDKVL 180
181 VIRVGYKYKC FAEDAVTVSR ILHIKLVPGK LTIDESNPQD CNHRQFAYCS FPDVRLNVHL 240
241 ERLVHHNLKV AVVEQAETSA IKKHDPGASK SSVFERKISN VFTKATFGVN STFVLRGKRI 300
301 LGDTNSIWAL SRDVHQGKVA KYSLISVNLN NGEVVYDEFE EPNLADEKLQ IRIKYLQPIE 360
361 VLVNTDDLPL HVAKFFKDIS CPLIHKQEYD LEDHVVQAIK VMNEKIQLSP SLIRLVSKLY 420
421 SHMVEYNNEQ VMLIPSIYSP FASKIHMLLD PNSLQSLDIF THDGGKGSLF WLLDHTRTSF 480
481 GLRMLREWIL KPLVDVHQIE ERLDAIECIT SEINNSIFFE SLNQMLNHTP DLLRTLNRIM 540
541 YGTTSRKEVY FYLKQITSFV DHFKMHQSYL SEHFKSSDGR IGKQSPLLFR LFSELNELLS 600
601 TTQLPHFLTM INVSAVMEKN SDKQVMDFFN LNNYDCSEGI IKIQRESESV RSQLKEELAE 660
661 IRKYLKRPYL NFRDEVDYLI EVKNSQIKDL PDDWIKVNNT KMVSRFTTPR TQKLTQKLEY 720
721 YKDLLIRESE LQYKEFLNKI TAEYTELRKI TLNLAQYDCI LSLAATSCNV NYVRPTFVNG 780
781 QQAIIAKNAR NPIIESLDVH YVPNDIMMSP ENGKINIITG PNMGGKSSYI RQVALLTIMA 840
841 QIGSFVPAEE IRLSIFENVL TRIGAHDDII NGDSTFKVEM LDILHILKNC NKRSLLLLDE 900
901 VGRGTGTHDG IAISYALIKY FSELSDCPLI LFTTHFPMLG EIKSPLIRNY HMDYVEEQKT 960
961 GEDWMSVIFL YKLKKGLTYN SYGMNVAKLA RLDKDIINRA FSISEELRKE SINEDALKLF1020
1021 SSLKRILKSD NITATDKLAK LLSLDIH |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
MSH2 MSH3 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
MSH2 MSH3 PMS1 |
| View Details | Gavin AC, et al. (2002) |

|
CMR1 DNA2 HDA1 HDA2 HDA3 HSC82 KAP95 LCD1 MEC1 MGM101 MLH1 MPH1 MSH2 MSH3 MSH6 OLA1 PGK1 PMS1 PRO2 RAD16 RAD52 RFA1 RFA2 RFA3 RIM1 RNH1 RSC8 RVB2 SGS1 SHS1 TIF1, TIF2 TOP3 VAS1 VPS1 YHR122W |
| View Details | Ho Y, et al. (2002) |

|
BIO3 HTB1 MSH3 |
| View Details | Qiu et al. (2008) |

|
MSH3 MSH6 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
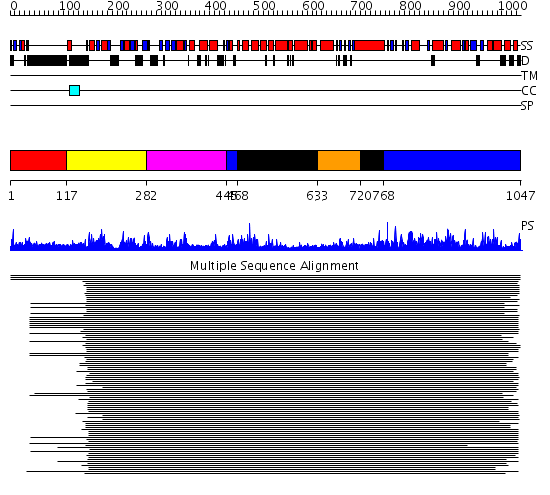
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [117..281] | 1510.457575 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I | |
| 3 | View Details | [282..444] | 1510.457575 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I | |
| 4 | View Details | [445..467] [768..1047] |
1510.457575 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I | |
| 5 | View Details | [468..632] [720..767] |
1510.457575 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I | |
| 6 | View Details | [633..719] | 1510.457575 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.39 |
Source: Reynolds et al. (2008)