













| General Information: |
|
| Name(s) found: |
RAD16 /
YBR114W
[SGD]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 790 amino acids |
Gene Ontology: |
|
| Cellular Component: |
repairosome
[IDA nucleotide-excision repair factor 4 complex [IDA |
| Biological Process: |
nucleotide-excision repair, DNA damage recognition
[IDA |
| Molecular Function: |
DNA-dependent ATPase activity
[IDA damaged DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQEGGFIRRR RTRSTKKSVN YNELSDDDTA VKNSKTLQLK GNSENVNDSQ DEEYRDDATL 60
61 VKSPDDDDKD FIIDLTGSDK ERTATDENTH AIKNDNDEII EIKEERDVSD DDEPLTKKRK 120
121 TTARKKKKKT STKKKSPKVT PYERNTLRLY EHHPELRNVF TDLKNAPPYV PQRSKQPDGM 180
181 TIKLLPFQLE GLHWLISQEE SIYAGGVLAD EMGMGKTIQT IALLMNDLTK SPSLVVAPTV 240
241 ALMQWKNEIE QHTKGQLKIY IYHGASRTTD IKDLQGYDVV LTTYAVLESV FRKQNYGFRR 300
301 KNGLFKQPSV LHNIDFYRVI LDEAHNIKDR QSNTARAVNN LKTQKRWCLS GTPLQNRIGE 360
361 MYSLIRFLNI NPFTKYFCTK CDCASKDWKF TDRMHCDHCS HVIMQHTNFF NHFMLKNIQK 420
421 FGVEGPGLES FNNIQTLLKN IMLRRTKVER ADDLGLPPRI VTVRRDFFNE EEKDLYRSLY 480
481 TDSKRKYNSF VEEGVVLNNY ANIFTLITRM RQLADHPDLV LKRLNNFPGD DIGVVICQLC 540
541 NDEAEEPIES KCHHKFCRLC IKEYVESFME NNNKLTCPVC HIGLSIDLSQ PALEVDLDSF 600
601 KKQSIVSRLN MSGKWQSSTK IEALVEELYK LRSNKRTIKS IVFSQFTSML DLVEWRLKRA 660
661 GFQTVKLQGS MSPTQRDETI KYFMNNIQCE VFLVSLKAGG VALNLCEASQ VFILDPWWNP 720
721 SVEWQSGDRV HRIGQYRPVK ITRFCIEDSI EARIIELQEK KANMIHATIN QDEAAISRLT 780
781 PADLQFLFNN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
RAD16 RAD7 |
| View Details | Gavin AC, et al. (2002) |
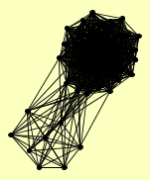
|
ADH1 CMR1 DNA2 HDA1 HDA2 HDA3 HSC82 KAP95 LCD1 MEC1 MGM101 MIR1 MLH1 MPH1 MSH2 MSH3 MSH6 OLA1 PDC1 PEP4 PGK1 PMS1 PRO2 RAD16 RAD52 RFA1 RFA2 RFA3 RIM1 RNH1 RSC8 RVB2 SGS1 SHS1 TDH3 TIF1, TIF2 TOP3 VAS1 VMA4 VPS1 WRS1 YHR122W YMR315W |
| View Details | Ho Y, et al. (2002) |
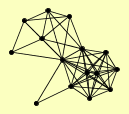
|
CCE1 CTF4 GND1 HHF1, HHF2 HTB2 HTZ1 PDX1 RAD1 RAD14 RAD16 RAD4 RAD7 SHP1 TMA29 TSA2 |
| View Details | Qiu et al. (2008) |
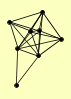
|
ABF1 ELC1 RAD14 RAD16 RAD2 RAD23 RAD4 RAD7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
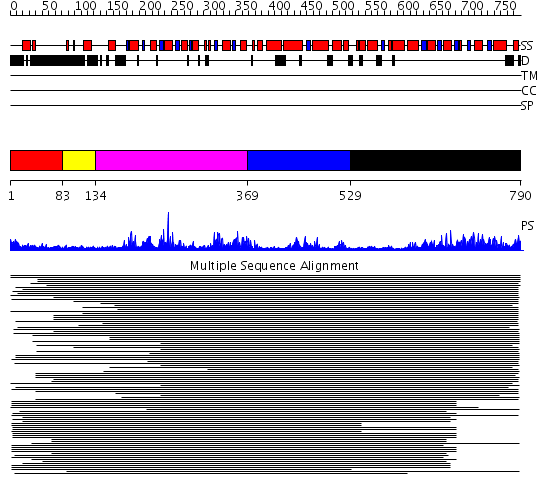
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [83..133] | 2.39794 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 3 | View Details | [134..368] | 2.39794 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 4 | View Details | [369..528] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [529..790] | 13.522879 | Putative DEAD box RNA helicase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)