













| General Information: |
|
| Name(s) found: |
RAD1 /
YPL022W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1100 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleotide-excision repair factor 1 complex
[IPI nucleus [IDA |
| Biological Process: |
nucleotide-excision repair, DNA incision, 5'-to lesion
[IDA double-strand break repair via single-strand annealing, removal of nonhomologous ends [IMP removal of nonhomologous ends [IMP meiotic mismatch repair [IMP mitotic recombination [IMP |
| Molecular Function: |
DNA binding
[IDA single-stranded DNA specific endodeoxyribonuclease activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQLFYQGDS DDELQEELTR QTTQASQSSK IKNEDEPDDS NHLNEVENED SKVLDDDAVL 60
61 YPLIPNEPDD IETSKPNIND IRPVDIQLTL PLPFQQKVVE NSLITEDALI IMGKGLGLLD 120
121 IVANLLHVLA TPTSINGQLK RALVLVLNAK PIDNVRIKEA LEELSWFSNT GKDDDDTAVE 180
181 SDDELFERPF NVVTADSLSI EKRRKLYISG GILSITSRIL IVDLLSGIVH PNRVTGMLVL 240
241 NADSLRHNSN ESFILEIYRS KNTWGFIKAF SEAPETFVME FSPLRTKMKE LRLKNVLLWP 300
301 RFRVEVSSCL NATNKTSHNK VIEVKVSLTN SMSQIQFGLM ECLKKCIAEL SRKNPELALD 360
361 WWNMENVLDI NFIRSIDSVM VPNWHRISYE SKQLVKDIRF LRHLLKMLVT SDAVDFFGEI 420
421 QLSLDANKPS VSRKYSESPW LLVDEAQLVI SYAKKRIFYK NEYTLEENPK WEQLIHILHD 480
481 ISHERMTNHL QGPTLVACSD NLTCLELAKV LNASNKKRGV RQVLLNKLKW YRKQREETKK 540
541 LVKEVQSQDT FPENATLNVS STFSKEQVTT KRRRTRGASQ VAAVEKLRNA GTNVDMEVVF 600
601 EDHKLSEEIK KGSGDDLDDG QEENAANDSK IFEIQEQENE ILIDDGDAEF DNGELEYVGD 660
661 LPQHITTHFN KDLWAEHCNE YEYVDRQDEI LISTFKSLND NCSLQEMMPS YIIMFEPDIS 720
721 FIRQIEVYKA IVKDLQPKVY FMYYGESIEE QSHLTAIKRE KDAFTKLIRE NANLSHHFET 780
781 NEDLSHYKNL AERKLKLSKL RKSNTRNAGG QQGFHNLTQD VVIVDTREFN ASLPGLLYRY 840
841 GIRVIPCMLT VGDYVITPDI CLERKSISDL IGSLQNNRLA NQCKKMLKYY AYPTLLIEFD 900
901 EGQSFSLEPF SERRNYKNKD ISTVHPISSK LSQDEIQLKL AKLVLRFPTL KIIWSSSPLQ 960
961 TVNIILELKL GREQPDPSNA VILGTNKVRS DFNSTAKGLK DGDNESKFKR LLNVPGVSKI1020
1021 DYFNLRKKIK SFNKLQKLSW NEINELINDE DLTDRIYYFL RTEKEEQEQE STDENLESPG1080
1081 KTTDDNALHD HHNDVPEAPV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
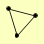
|
RAD1 RAD10 SAW1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
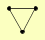
|
RAD1 RAD10 RAD14 |
| View Details | Krogan NJ, et al. (2006) |
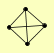
|
DPS1 RAD1 RAD10 SAW1 |
| View Details | Gavin AC, et al. (2002) |
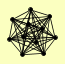
|
ADH1 GIN4 HMO1 KCC4 PBP2 PDC1 PGI1 RAD1 RAD10 YEF3 YRA1 |
| View Details | Ho Y, et al. (2002) |
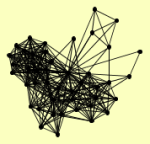
|
ARC1 CAR2 CDC60 CPR1 CTF4 DEF1 DMA2 DUN1 FAA1 FAR1 FUM1 GDE1 GPD1 GPD2 HEF3 KGD1 LSP1 MDH1 MDM30 MSI1 PDC6 PIL1 PRO1 PRO2 PTC6 PYC1 RAD1 RAD10 RAD14 RAD16 RAD4 RNR2 SAH1 SAW1 SEC6 SEN1 SOD2 STE20 TFP1 TIF1, TIF2 TPS1 UBI4 UTP4 YHR033W |
| View Details | Qiu et al. (2008) |
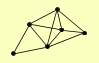
|
ABF1 RAD1 RAD10 RAD14 RAD2 RAD7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
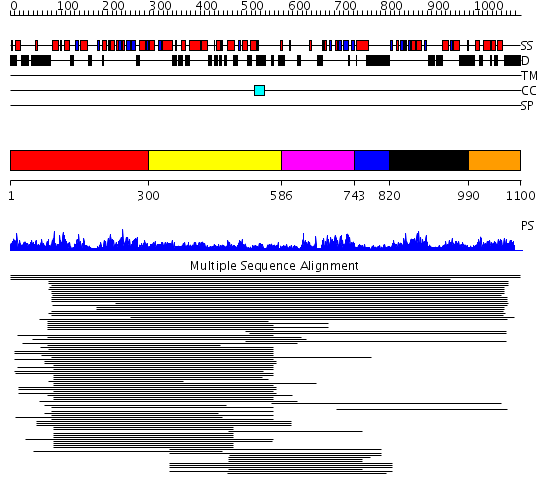
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..299] | 12.522879 | Initiation factor 4a | |
| 2 | View Details | [300..585] | 3.0 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 3 | View Details | [586..742] | 3.0 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 4 | View Details | [743..819] | 28.0 | Nucleotide excision repair enzyme UvrB | |
| 5 | View Details | [820..989] | 113.28043 | Putative ATP-dependent RNA helicase Hef, nuclease domain | |
| 6 | View Details | [990..1100] | 2.057993 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)