













| General Information: |
|
| Name(s) found: |
YRA1 /
YDR381W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 226 amino acids |
Gene Ontology: |
|
| Cellular Component: |
transcription export complex
[IPI |
| Biological Process: |
mRNA export from nucleus
[IGI |
| Molecular Function: |
RNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSANLDKSLD EIIGSNKAGS NRARVGGTRG NGPRRVGKQV GSQRRSLPNR RGPIRKNTRA 60
61 PPNAVARVAK LLDTTREVKV NVEGLPRDIK QDAVREFFAS QVGGVQRVLL SYNERGQSTG 120
121 MANITFKNGE LARRAVERFN GSPIDGGRSR LRLNLIVDPN QRPVKSLADR IKAMPQKGGN 180
181 APRPVKRGPN RKAAMAKSQN KPKREKPAKK SLEDLDKEMA DYFEKK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
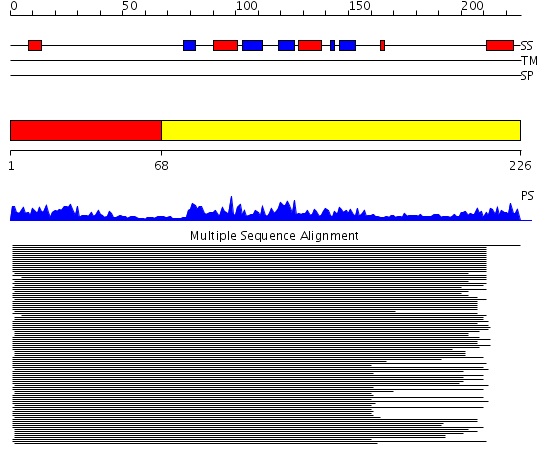
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..67] | 46.154902 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) | |
| 2 | View Details | [68..226] | 46.154902 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)