













| General Information: |
|
| Name(s) found: |
HOS4 /
YIL112W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1083 amino acids |
Gene Ontology: |
|
| Cellular Component: |
histone deacetylase complex
[IPI |
| Biological Process: |
histone deacetylation
[IDA negative regulation of meiosis [IPI |
| Molecular Function: |
NAD-dependent histone deacetylase activity
[IDA NAD-independent histone deacetylase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNETTTKQPL KKRSLSSYLS NVSTRREELE KISKQETSEE EDTAGKHEQR ETLSEEVSDK 60
61 FPENVASFRS QTTSVHQATQ NNLNAKESED LAHKNDASSH EGEVNGDSRP DDVPETNEKI 120
121 SQAIRAKISS SSSSPNVRNV DIQNHQPFSR DQLRAMLKEP KRKTVDDFIE EEGLGAVEEE 180
181 DLSDEVLEKN TTEPENVEKD IEYSDSDKDT DDVGSDDPTA PNSPIKLGRR KLVRGDQLDA 240
241 TTSSMFNNES DSELSDIDDS KNIALSSSLF RGGSSPVKET NNNLSNMNSS PAQNPKRGSV 300
301 SRSNDSNKSS HIAVSKRPKQ KKGIYRDSGG RTRLQIACDK GKYDVVKKMI EEGGYDINDQ 360
361 DNAGNTALHE AALQGHIEIV ELLIENGADV NIKSIEMFGD TPLIDASANG HLDVVKYLLK 420
421 NGADPTIRNA KGLTAFESVD DESEFDDEED QKILREIKKR LSIAAKKWTN RAGIHNDKSK 480
481 NGNNAHTIDQ PPFDNTTKAK NEKAADSPSM ASNIDEKAPE EEFYWTDVTS RAGKEKLFKA 540
541 SKEGHLPYVG TYVENGGKID LRSFFESVKC GHEDITSIFL AFGFPVNQTS RDNKTSALMV 600
601 AVGRGHLGTV KLLLEAGADP TKRDKKGRTA LYYAKNSIMG ITNSEEIQLI ENAINNYLKK 660
661 HSEDNNDDDD DDDNNNETYK HEKKREKTQS PILASRRSAT PRIEDEEDDT RMLNLADDDF 720
721 NNDRDVKEST TSDSRKRLDD NENVGTQYSL DWKKRKTNAL QDEEKLKSIS PLSMEPHSPK 780
781 KAKSVEISKI HEETAAEREA RLKEEEEYRK KRLEKKRKKE QELLQKLAED EKKRIEEQEK 840
841 QKVLEMERLE KATLEKARKM EREKEMEEIS YRRAVRDLYP LGLKIINFND KLDYKRFLPL 900
901 YYFVDEKNDK FVLDLQVMIL LKDIDLLSKD NQPTSEKIPV DPSHLTPLWN MLKFIFLYGG 960
961 SYDDKKNNME NKRYVVNFDG VDLDTKIGYE LLEYKKFVSL PMAWIKWDNV VIENHAKRKE1020
1021 IEGNMIQISI NEFARWRNDK LNKAQQPTRK QRSLKIPREL PVKFQHRMSI SSVLQQTSKE1080
1081 PFW |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CPR1 HOS2 HOS4 SET3 SIF2 SNT1 |
| View Details | Krogan NJ, et al. (2006) |
|
AKL1 HOS4 HST1 RFM1 SET3 SIF2 SNT1 SUM1 |
| View Details | Gavin AC, et al. (2006) |
|
CPR1 HOS2 HOS4 SET3 SIF2 SNT1 |
| View Details | Gavin AC, et al. (2002) |
|
BLM10 CDC55 CIN1 CPR1 ERG13 GUS1 HHF1, HHF2 HOS2 HOS4 IML1 KAP95 KEL1 LTE1 MYO5 PFK1 PPH21 PPH22 PRE1 PRE10 PRE2 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 RRD2 RTS1 RTS3 RVB2 SCL1 SET3 SIF2 SNF2 SNT1 SRP1 TDH2 TDH3 TEF4 TPD3 YBL104C YOR1 YRA1 ZDS1 ZDS2 |
| View Details | Ho Y, et al. (2002) |
|
AVT2 HOS4 SIF2 SNT1 TFP1 TRM3 UTP21 YMR155W YRF1-3, YRF1-7 ZDS2 |
| View Details | Qiu et al. (2008) |
|
HDA1 HDA2 HDA3 HOS1 HOS2 HOS4 HST1 RPD3 SDS3 SET3 SIF2 SNT1 STB2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
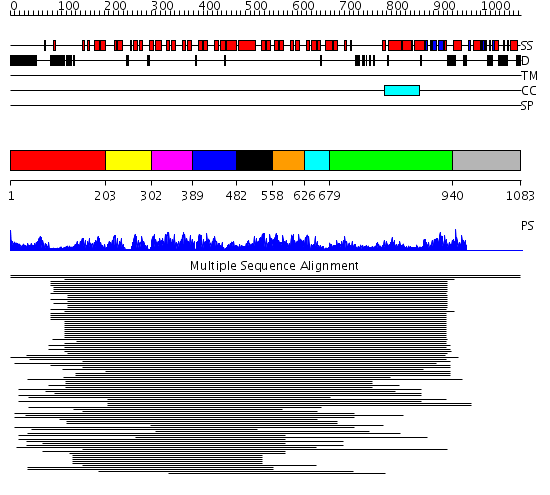
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..202] | 2.237994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [203..301] | 73.67837 | p18ink4C(ink6) | |
| 3 | View Details | [302..388] | 73.67837 | p18ink4C(ink6) | |
| 4 | View Details | [389..481] | 111.9897 | GA bindinig protein (GABP) beta 1 | |
| 5 | View Details | [482..557] | 3.516997 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [558..625] | 134.29073 | Myotrophin | |
| 7 | View Details | [626..678] | 134.29073 | Myotrophin | |
| 8 | View Details | [679..939] | 11.83 | No description for 1ldvA was found. | |
| 9 | View Details | [940..1083] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)