













| General Information: |
|
| Name(s) found: |
HST1 /
YOL068C
[SGD]
|
| Description(s) found:
Found 34 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 503 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS histone deacetylase complex [IDA |
| Biological Process: |
telomere maintenance
[IMP chromatin silencing [IGI negative regulation of mitotic recombination [IMP |
| Molecular Function: |
NAD-dependent histone deacetylase activity
[IDA NAD-independent histone deacetylase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNILLMQRIV SFILVVSQGR YFHVGELTMT MLKRPQEEES DNNATKKLKT RLTYPCILGK 60
61 DKVTGKFIFP AITKDDVMNA RLFLKDNDLK TFLEYFLPVE VNSIYIYFMI KLLGFDVKDK 120
121 ELFMALNSNI TSNKERSSAE LSSIHAKAED EDELTDPLEK KHAVKLIKDL QKAINKVLST 180
181 RLRLPNFNTI DHFTATLRNA KKILVLTGAG VSTSLGIPDF RSSEGFYSKI RHLGLEDPQD 240
241 VFNLDIFLQD PSVFYNIAHM VLPPENMYSP LHSFIKMLQD KGKLLRNYTQ NIDNLESYAG 300
301 IDPDKLVQCH GSFATASCVT CHWQIPGEKI FENIRNLELP LCPYCYQKRK QYFPMSNGNN 360
361 TVQTNINFNS PILKSYGVLK PDMTFFGEAL PSRFHKTIRK DILECDLLIC IGTSLKVAPV 420
421 SEIVNMVPSH VPQILINRDM VTHAEFDLNL LGFCDDVASL VAKKCHWDIP HKKWQDLKKI 480
481 DYNCTEIDKG TYKIKKQPRK KQQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
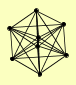
|
AKL1 HOS4 HST1 RFM1 SET3 SIF2 SNT1 SUM1 |
| View Details | Gavin AC, et al. (2002) |

|
HST1 SIF2 |
| View Details | Qiu et al. (2008) |
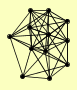
|
HDA1 HDA3 HOS1 HOS4 HST1 PHO23 RXT2 SAP30 SDS3 SET3 SIF2 SNT1 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
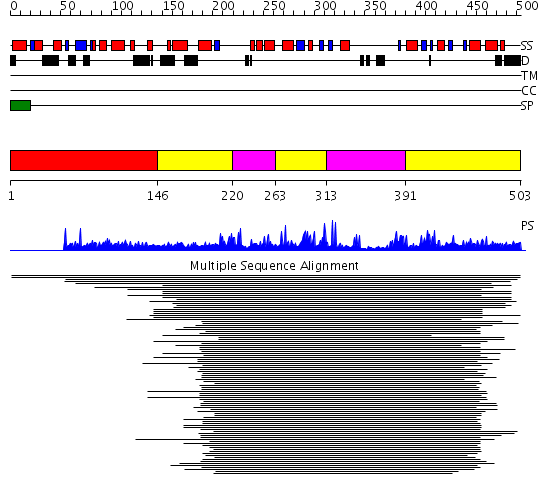
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..145] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [146..219] [263..312] [391..503] |
475.39794 | Sirt2 histone deacetylase | |
| 3 | View Details | [220..262] [313..390] |
475.39794 | Sirt2 histone deacetylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.59 |
Source: Reynolds et al. (2008)