













| General Information: |
|
| Name(s) found: |
NOP56 /
YLR197W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 504 amino acids |
Gene Ontology: |
|
| Cellular Component: |
box C/D snoRNP complex
[IDA nucleus [IDA nuclear outer membrane [TAS nucleolus [IDA small nucleolar ribonucleoprotein complex [IPI |
| Biological Process: |
rRNA modification
[TAS maturation of SSU-rRNA [IPI ribosome biogenesis [RCA |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPIEYLLFE EPTGYAVFKV KLQQDDIGSR LKEVQEQIND FGAFTKLIEL VSFAPFKGAA 60
61 EALENANDIS EGLVSESLKA ILDLNLPKAS SKKKNITLAI SDKNLGPSIK EEFPYVDCIS 120
121 NELAQDLIRG VRLHGEKLFK GLQSGDLERA QLGLGHAYSR AKVKFSVQKN DNHIIQAIAL 180
181 LDQLDKDINT FAMRVKEWYG WHFPELAKLV PDNYTFAKLV LFIKDKASLN DDSLHDLAAL 240
241 LNEDSGIAQR VIDNARISMG QDISETDMEN VCVFAQRVAS LADYRRQLYD YLCEKMHTVA 300
301 PNLSELIGEV IGARLISHAG SLTNLSKQAA STVQILGAEK ALFRALKTKG NTPKYGLIYH 360
361 SGFISKASAK NKGRISRYLA NKCSMASRID NYSEEPSNVF GSVLKKQVEQ RLEFYNTGKP 420
421 TLKNELAIQE AMELYNKDKP AAEVEETKEK ESSKKRKLED DDEEKKEKKE KKSKKEKKEK 480
481 KEKKDKKEKK DKKEKKDKKK KSKD |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | control 1 | McCusker D, et al (2007) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | Boi 2 gst control | McCusker D, et al (2007) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
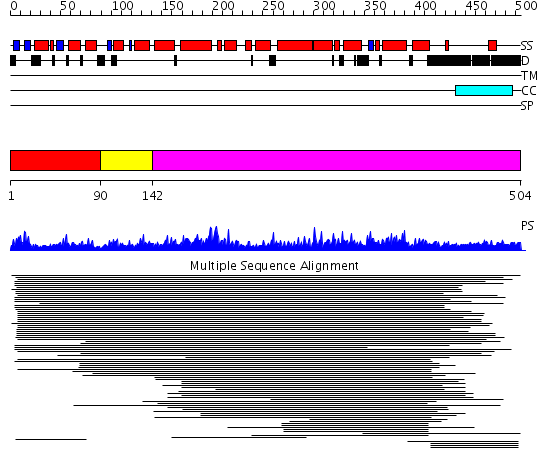
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | 1.037982 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [90..141] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [142..504] | 14.062961 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)