













| General Information: |
|
| Name(s) found: |
YGR283C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 341 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IDA ribosome [IDA |
| Biological Process: |
ribosome biogenesis
[RCA |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAVKHKSESL KHEEGAAKKA KTGLLKLKKI MDIESNVVKY SICIPTTVID NCNNLEQVTF 60
61 TAYQIARTAV LFNVQEIIVL DQSKDKKHEK KSRSKETISD CLLLATLLQY FVTPPNLLDT 120
121 TFKKKNKLYL KCASTFPPLN QLPFMNASAE QHYKEGLSIA RDSSKGKSDD ALTNLVYIGK 180
181 NQIITLSNQN IPNTARVTVD TERKEVVSPI DAYKGKPLGY HVRMASTLNE VSEGYTKIVW 240
241 VNSGDFHYDE ELSKYHKVET KLPYIAKLKK SSTSEKPCNI LLIFGKWGHL KRCFRRSDLE 300
301 SSSLHHYFSG QLQFPASVPQ GNIPIQDSLP IALTMFQRWA S |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
GAR1 NHP2 YGR283C YMR310C |
| View Details | Krogan NJ, et al. (2006) |
|
SAN1 YGR283C YMR310C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
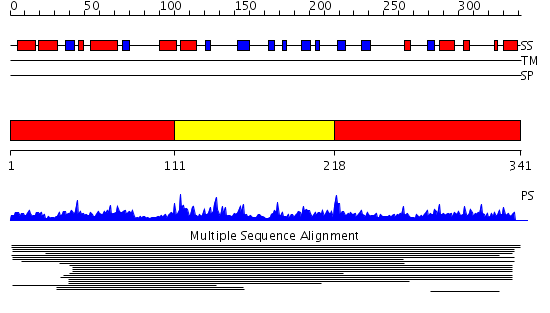
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..110] [218..341] |
47.39794 | Hypothetical protein MTH1 (MT0001), insert domain; Hypothetical protein MTH1 (MT0001), dimerisation domain | |
| 2 | View Details | [111..217] | 47.39794 | Hypothetical protein MTH1 (MT0001), insert domain; Hypothetical protein MTH1 (MT0001), dimerisation domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)