













| General Information: |
|
| Name(s) found: |
DBP3 /
YGL078C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 523 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear outer membrane
[IMP nucleolus [IDA |
| Biological Process: |
ribosomal large subunit assembly
[IMP |
| Molecular Function: |
ATP-dependent RNA helicase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTKEEIADKK RKVVDEEVIE KKKSKKHKKD KKDKKEKKDK KHKKHKKEKK GEKEVEVPEK 60
61 ESEKKPEPTS AVASEFYVQS EALTSLPQSD IDEYFKENEI AVEDSLDLAL RPLLSFDYLS 120
121 LDSSIQAEIS KFPKPTPIQA VAWPYLLSGK DVVGVAETGS GKTFAFGVPA ISHLMNDQKK 180
181 RGIQVLVISP TRELASQIYD NLIVLTDKVG MQCCCVYGGV PKDEQRIQLK KSQVVVATPG 240
241 RLLDLLQEGS VDLSQVNYLV LDEADRMLEK GFEEDIKNII RETDASKRQT LMFTATWPKE 300
301 VRELASTFMN NPIKVSIGNT DQLTANKRIT QIVEVVDPRG KERKLLELLK KYHSGPKKNE 360
361 KVLIFALYKK EAARVERNLK YNGYNVAAIH GDLSQQQRTQ ALNEFKSGKS NLLLATDVAA 420
421 RGLDIPNVKT VINLTFPLTV EDYVHRIGRT GRAGQTGTAH TLFTEQEKHL AGGLVNVLNG 480
481 ANQPVPEDLI KFGTHTKKKE HSAYGSFFKD VDLTKKPKKI TFD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
COQ10 DBP3 DSE4 GAR1 KRI1 KRR1 NHP2 NOP10 NOP6 YCR016W YDR381C-A |
| View Details | Qiu et al. (2008) |
|
BFR2 BRX1 DBP10 DBP2 DBP3 DBP6 DBP7 DBP8 DBP9 DHR2 DRS1 ECM16 ERB1 GAR1 HAS1 HCA4 KRI1 KRR1 MAK5 MTR4 NOC3 NOG2 NOP12 NOP13 NOP15 NOP4 NOP7 NSR1 NUG1 PRP43 PXR1 RNT1 ROK1 RPF2 RRP1 RRP3 RRP8 SOF1 SPB4 UTP10 UTP13 UTP15 UTP4 UTP5 UTP6 UTP8 UTP9 YBL028C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
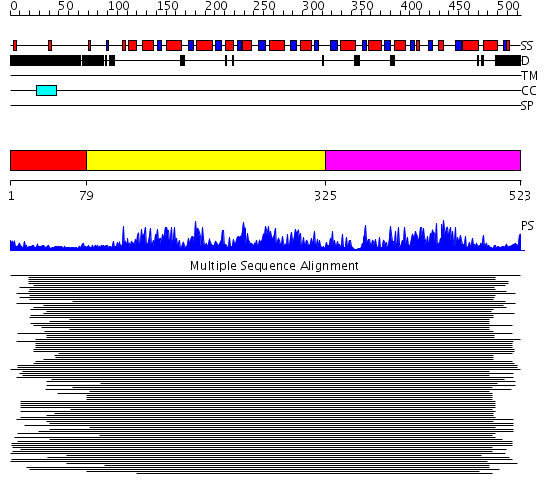
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..78] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [79..324] | 636.9794 | Putative DEAD box RNA helicase | |
| 3 | View Details | [325..523] | 636.9794 | Putative DEAD box RNA helicase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)