













| General Information: |
|
| Name(s) found: |
DBP9 /
YLR276C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 594 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear outer membrane
[IMP nucleolus [TAS |
| Biological Process: |
ribosome biogenesis
[RCA ribosomal large subunit assembly [IMP |
| Molecular Function: |
ATP-dependent RNA helicase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSYEKKSVEG AYIDDSTTFE AFHLDSRLLQ AIKNIGFQYP TLIQSHAIPL ALQQKRDIIA 60
61 KAATGSGKTL AYLIPVIETI LEYKKTIDNG EENGTLGIIL VPTRELAQQV YNVLEKLVLY 120
121 CSKDIRTLNI SSDMSDSVLS TLLMDQPEII VGTPGKLLDL LQTKINSISL NELKFLVVDE 180
181 VDLVLTFGYQ DDLNKIGEYL PLKKNLQTFL MSATLNDDIQ ALKQKFCRSP AILKFNDEEI 240
241 NKNQNKLLQY YVKVSEFDKF LLCYVIFKLN LIKGKTLIFV NNIDRGYRLK LVMEQFGIKS 300
301 CILNSELPVN SRQHIVDQFN KNVYQLLIAT DDTEYIKEED DEIEEGHNTE NQEEKSLEGE 360
361 PENDKKPSKK KKVQVKKDKE YGVSRGVDFK NVACVLNFDL PTTAKSYVHR VGRTARGGKT 420
421 GTAISFVVPL KEFGKHKPSM LQTAKKDERI LSRIIKQQSK LGLELQPYKF DQKQVEAFRY 480
481 RMEDGFRAVT QVAIREARVK ELKQELLASE KLKRHFEENP KELQSLRHDK ELHPARVQQH 540
541 LKRVPDYLLP ESARGNGTKV KFVPFHNAKK RHSHKKGRVS KPKNGKVDPL KNFK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
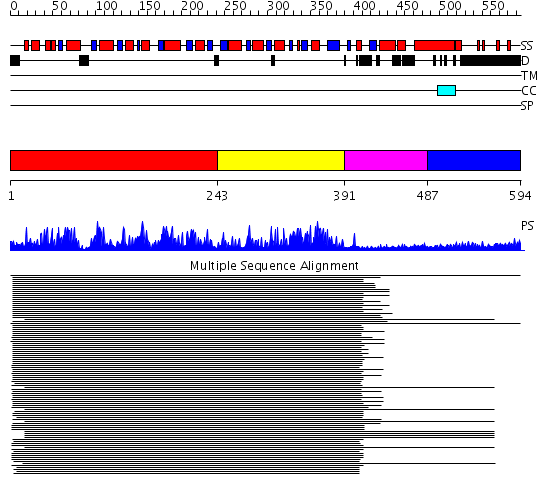
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..242] | 394.9897 | Putative DEAD box RNA helicase | |
| 2 | View Details | [243..390] | 394.9897 | Putative DEAD box RNA helicase | |
| 3 | View Details | [391..486] | 9.154902 | Nucleotide excision repair enzyme UvrB | |
| 4 | View Details | [487..594] | 1.168988 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)