













| General Information: |
|
| Name(s) found: |
NOP2 /
YNL061W
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 618 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[TAS |
| Biological Process: |
ribosome biogenesis
[RCA rRNA processing [TAS |
| Molecular Function: |
RNA methyltransferase activity
[TAS S-adenosylmethionine-dependent methyltransferase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSRRHKNKQ AAPPTLEEFQ ARKEKKANRK LEKGKRPSTT QGDEVSDRKK KKSKPFKKSR 60
61 KEEEEVVEED KDLPEVDLEE LSKARKSLFD DEEDDDEAGL VDEELKDEFD LEQEYDYDED 120
121 EDNDAHPIFS DDDDEADLEE LNAQNMEALS KKLDEEEAEE AEEAEMELVE AENMQPRADI 180
181 LPTEEQEEMM AQETPNLTST RTRMIEIVKV LENFKTLGAE GRSRGEYVDR LLKDICEYFG 240
241 YTPFLAEKLF NLFSPAEAME FFEANEIARP ITIRTNTLKT RRRDLAQTLV NRGVNLQPIG 300
301 SWTKVGLQIF DSQVPIGATP EYLAGHYILQ AASSFLPVIA LDPHENERIL DMAAAPGGKT 360
361 TYISAMMKNT GCVFANDANK SRTKSLIANI HRLGCTNTIV CNYDAREFPK VIGGFDRILL 420
421 DAPCSGTGVI GKDQSVKVSR TEKDFIQIPH LQKQLLLSAI DSVDCNSKHG GVIVYSTCSV 480
481 AVEEDEAVID YALRKRPNVK LVDTGLAIGK EAFTSYRGKK FHPSVKLARR YYPHTYNVDG 540
541 FFVAKFQKIG PSSFDDNQAS AKEKETAARK EALEEGIIHS DFATFEDEED DKYIEKSVKN 600
601 NLLKKGVNPK AKRPSNEK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
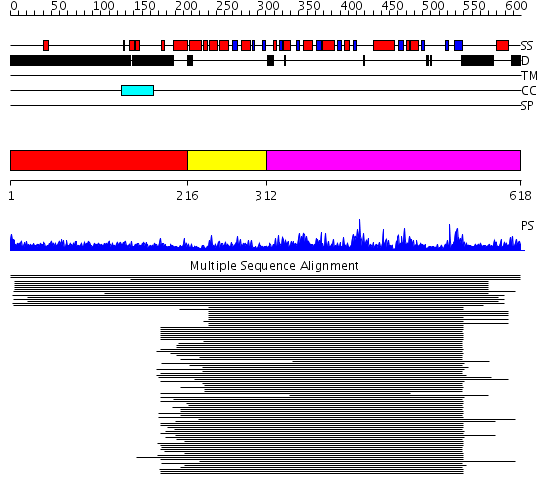
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..215] | 28.071997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [216..311] | 3.163984 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [312..618] | 270.917458 | No description for 1j4fA_ was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)