













| General Information: |
|
| Name(s) found: |
NOB1 /
YOR056C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 459 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA preribosome [IDA |
| Biological Process: |
maturation of SSU-rRNA
[IEP ribosomal small subunit biogenesis [IPI ribosome biogenesis [RCA proteasome assembly [IMP |
| Molecular Function: |
RNA binding
[IDA protein binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTENQTAHVR ALILDATPLI TQSYTHYQNY AQSFYTTPTV FQEIKDAQAR KNLEIWQSLG 60
61 TLKLVHPSEN SIAKVSTFAK LTGDYSVLSA NDLHILALTY ELEIKLNNGD WRLRKKPGDA 120
121 LDASKADVGT DGKQKLTEDN KKEEDSESVP KKKNKRRGGK KQKAKREARE AREAENANLE 180
181 LESKAEEHVE EAGSKEQICN DENIKESSDL NEVFEDADDD GDWITPENLT EAIIKDSGED 240
241 TTGSLGVEAS EEDRHVALNR PENQVALATG DFAVQNVALQ MNLNLMNFMS GLKIKRIRNY 300
301 MLRCHACFKI FPLPKDGKPK HFCASCGGQG TLLRCAVSVD SRTGNVTPHL KSNFQWNNRG 360
361 NRYSVASPLS KNSQKRYGKK GHVHSKPQEN VILREDQKEY EKVIKQEEWT RRHNEKILNN 420
421 WIGGGSADNY ISPFAITGLK QHNVRIGKGR YVNSSKRRS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Unpublished Data |
| View Data |
|
Huh WK, et al. (2003) |
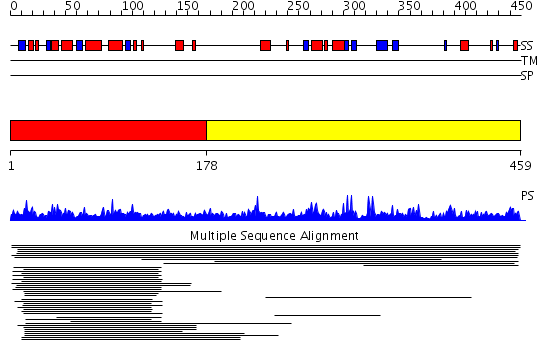
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..177] | 15.041993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [178..459] | 1.015926 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [223..386] | 23.0 | Solution Structure of RSGI RUH-035, a Zn-ribbon module in Mouse cDNA | |
| 4 | View Details | [387..459] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)