













| General Information: |
|
| Name(s) found: |
ENP1 /
YBR247C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 483 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear outer membrane [IMP nucleolus [TAS preribosome, small subunit precursor [IDA |
| Biological Process: |
ribosome biogenesis
[RCA rRNA processing [IPI |
| Molecular Function: |
snoRNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARASSTKAR KQRHDPLLKD LDAAQGTLKK INKKKLAQND AANHDAANEE DGYIDSKASR 60
61 KILQLAKEQQ DEIEGEELAE SERNKQFEAR FTTMSYDDED EDEDEDEEAF GEDISDFEPE 120
121 GDYKEEEEIV EIDEEDAAMF EQYFKKSDDF NSLSGSYNLA DKIMASIREK ESQVEDMQDD 180
181 EPLANEQNTS RGNISSGLKS GEGVALPEKV IKAYTTVGSI LKTWTHGKLP KLFKVIPSLR 240
241 NWQDVIYVTN PEEWSPHVVY EATKLFVSNL TAKESQKFIN LILLERFRDN IETSEDHSLN 300
301 YHIYRAVKKS LYKPSAFFKG FLFPLVETGC NVREATIAGS VLAKVSVPAL HSSAALSYLL 360
361 RLPFSPPTTV FIKILLDKKY ALPYQTVDDC VYYFMRFRIL DDGSNGEDAT RVLPVIWHKA 420
421 FLTFAQRYKN DITQDQRDFL LETVRQRGHK DIGPEIRREL LAGASREFVD PQEANDDLMI 480
481 DVN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
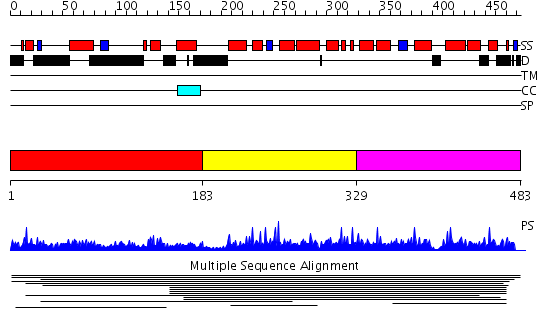
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..182] | 1.008981 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [183..328] | 1.008977 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [329..483] | 1.007957 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)