













| General Information: |
|
| Name(s) found: |
RAD50 /
YNL250W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1312 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrion [IDA Mre11 complex [IPI |
| Biological Process: |
meiotic DNA double-strand break processing
[TAS double-strand break repair via break-induced replication [TAS telomere maintenance [IMP telomere maintenance via recombination [IMP double-strand break repair via nonhomologous end joining [IMP meiosis [IMP meiotic DNA double-strand break formation [TAS |
| Molecular Function: |
ATPase activity
[IDA adenylate kinase activity [IDA protein binding [IPI telomeric DNA binding [IDA double-stranded DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAIYKLSIQ GIRSFDSNDR ETIEFGKPLT LIVGMNGSGK TTIIECLKYA TTGDLPPNSK 60
61 GGVFIHDPKI TGEKDIRAQV KLAFTSANGL NMIVTRNIQL LMKKTTTTFK TLEGQLVAIN 120
121 NSGDRSTLST RSLELDAQVP LYLGVPKAIL EYVIFCHQED SLWPLSEPSN LKKKFDEIFQ 180
181 AMKFTKALDN LKSIKKDMSV DIKLLKQSVE HLKLDKDRSK AMKLNIHQLQ TKIDQYNEEV 240
241 SEIESQLNEI TEKSDKLFKS NQDFQKILSK VENLKNTKLS ISDQVKRLSN SIDILDLSKP 300
301 DLQNLLANFS KVLMDKNNQL RDLETDISSL KDRQSSLQSL SNSLIRRQGE LEAGKETYEK 360
361 NRNHLSSLKE AFQHKFQGLS NIENSDMAQV NHEMSQFKAF ISQDLTDTID QFAKDIQLKE 420
421 TNLSDLIKSI TVDSQNLEYN KKDRSKLIHD SEELAEKLKS FKSLSTQDSL NHELENLKTY 480
481 KEKLQSWESE NIIPKLNQKI EEKNNEMIIL ENQIEKFQDR IMKTNQQADL YAKLGLIKKS 540
541 INTKLDELQK ITEKLQNDSR IRQVFPLTQE FQRADLEMDF QKLFINMQKN IAINNKKMHE 600
601 LDRRYTNALY NLNTIEKDLQ DNQKSKEKVI QLLSENLPED CTIDEYNDVL EETELSYKTA 660
661 LENLKMHQTT LEFNRKALEI AERDSCCYLC SRKFENESFK SKLLQELKTK TDANFEKTLK 720
721 DTVQNEKEYL HSLRLLEKHI ITLNSINEKI DNSQKCLEKA KEETKTSKSK LDELEVDSTK 780
781 LKDEKELAES EIRPLIEKFT YLEKELKDLE NSSKTISEEL SIYNTSEDGI QTVDELRDQQ 840
841 RKMNDSLREL RKTISDLQME KDEKVRENSR MINLIKEKEL TVSEIESSLT QKQNIDDSIR 900
901 SKRENINDID SRVKELEARI ISLKNKKDEA QSVLDKVKNE RDIQVRNKQK TVADINRLID 960
961 RFQTIYNEVV DFEAKGFDEL QTTIKELELN KAQMLELKEQ LDLKSNEVNE EKRKLADSNN1020
1021 EEKNLKQNLE LIELKSQLQH IESEISRLDV QNAEAERDKY QEESLRLRTR FEKLSSENAG1080
1081 KLGEMKQLQN QIDSLTHQLR TDYKDIEKNY HKEWVELQTR SFVTDDIDVY SKALDSAIMK1140
1141 YHGLKMQDIN RIIDELWKRT YSGTDIDTIK IRSDEVSSTV KGKSYNYRVV MYKQDVELDM1200
1201 RGRCSAGQKV LASIIIRLAL SETFGANCGV IALDEPTTNL DEENIESLAK SLHNIINMRR1260
1261 HQKNFQLIVI THDEKFLGHM NAAAFTDHFF KVKRDDRQKS QIEWVDINRV TY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
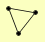
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
MRE11 RAD50 XRS2 |
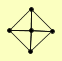
| View Details | Krogan NJ, et al. (2006) |
|
DMC1 MRE11 RAD50 XRS2 YBL044W |
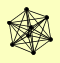
| View Details | Gavin AC, et al. (2002) |
|
ENP1 HRR25 PAA1 PFK1 PTC3 RAD50 TSR1 VPS15 |
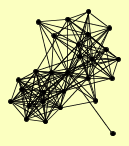
| View Details | Ho Y, et al. (2002) |
|
AHP1 CDC16 CDC39 DCS1 DCS2 DUN1 ERG20 GDB1 GPH1 MAM33 MKT1 MRE11 PST2 RAD50 REX2 RPT3 SEC27 SSK22 TFP1 TSA2 UBP15 UBR1 VMA8 VPS1 XRS2 YAK1 YLR241W YPL247C |
| View Details | Qiu et al. (2008) |

|
RAD50 XRS2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
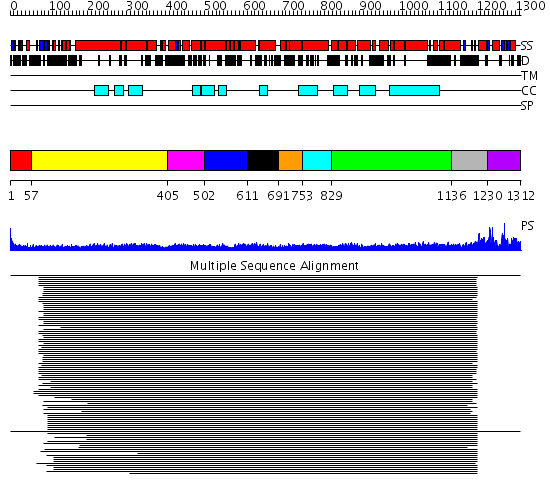
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..56] | 2.39794 | Rad50 | |
| 2 | View Details | [57..404] | 78.228787 | Heavy meromyosin subfragment | |
| 3 | View Details | [405..501] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [502..610] | 9.69897 | Tropomyosin | |
| 5 | View Details | [611..690] | 9.69897 | Tropomyosin | |
| 6 | View Details | [691..752] | 9.69897 | Tropomyosin | |
| 7 | View Details | [753..828] | 9.69897 | Tropomyosin | |
| 8 | View Details | [829..1135] | 33.337996 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1136..1229] | 38.440336 | CRYSTAL STRUCTURE OF ATP-FREE RAD50 ABC-ATPASE | |
| 10 | View Details | [1230..1312] | 38.440336 | CRYSTAL STRUCTURE OF ATP-FREE RAD50 ABC-ATPASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)