













| General Information: |
|
| Name(s) found: |
SEC27 /
YGL137W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 889 amino acids |
Gene Ontology: |
|
| Cellular Component: |
COPI vesicle coat
[IGI |
| Biological Process: |
ER to Golgi vesicle-mediated transport
[IMP retrograde vesicle-mediated transport, Golgi to ER [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLDIKKTFS NRSDRVKGID FHPTEPWVLT TLYSGRVELW NYETQVEVRS IQVTETPVRA 60
61 GKFIARKNWI IVGSDDFRIR VFNYNTGEKV VDFEAHPDYI RSIAVHPTKP YVLSGSDDLT 120
121 VKLWNWENNW ALEQTFEGHE HFVMCVAFNP KDPSTFASGC LDRTVKVWSL GQSTPNFTLT 180
181 TGQERGVNYV DYYPLPDKPY MITASDDLTI KIWDYQTKSC VATLEGHMSN VSFAVFHPTL 240
241 PIIISGSEDG TLKIWNSSTY KVEKTLNVGL ERSWCIATHP TGRKNYIASG FDNGFTVLSL 300
301 GNDEPTLSLD PVGKLVWSGG KNAAASDIFT AVIRGNEEVE QDEPLSLQTK ELGSVDVFPQ 360
361 SLAHSPNGRF VTVVGDGEYV IYTALAWRNK AFGKCQDFVW GPDSNSYALI DETGQIKYYK 420
421 NFKEVTSWSV PMHSAIDRLF SGALLGVKSD GFVYFFDWDN GTLVRRIDVN AKDVIWSDNG 480
481 ELVMIVNTNS NGDEASGYTL LFNKDAYLEA ANNGNIDDSE GVDEAFDVLY ELSESITSGK 540
541 WVGDVFIFTT ATNRLNYFVG GKTYNLAHYT KEMYLLGYLA RDNKVYLADR EVHVYGYEIS 600
601 LEVLEFQTLT LRGEIEEAIE NVLPNVEGKD SLTKIARFLE GQEYYEEALN ISPDQDQKFE 660
661 LALKVGQLTL ARDLLTDESA EMKWRALGDA SLQRFNFKLA VEAFTNAHDL ESLFLLHSSF 720
721 NNKEGLVTLA KDAERAGKFN LAFNAYWIAG DIQGAKDLLI KSQRFSEAAF LGSTYGLGDD 780
781 AVNDIVTKWK ENLILNGKNT VSERVCGAEG LPGSSSSGDA QPLIDLDSTP APEQADENKE 840
841 AEVEDSEFKE SNSEAVEAEK KEEEAPQQQQ SEQQPEQGEA VPEPVEEES |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
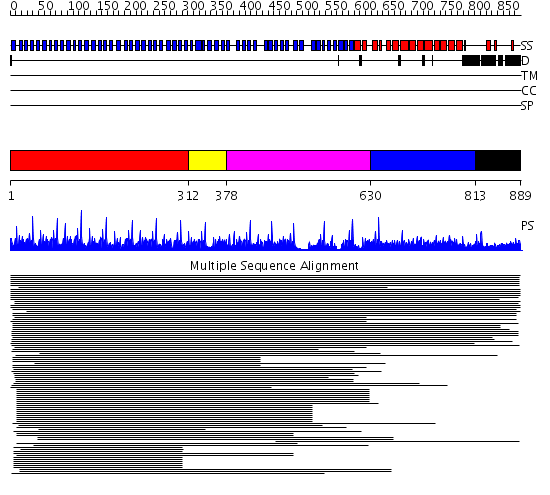
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..311] | 219.68867 | Tup1, C-terminal domain | |
| 2 | View Details | [312..377] | 3.106993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [378..629] | 3.0 | TolB, C-terminal domain; TolB, N-terminal domain | |
| 4 | View Details | [630..812] | 10.2 | Clathrin heavy chain proximal leg segment | |
| 5 | View Details | [813..889] | 1.023991 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)