













| General Information: |
|
| Name(s) found: |
NTG1 /
YAL015C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 399 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrion [IDA |
| Biological Process: |
DNA repair
[IDA base-excision repair [IDA base-excision repair, AP site formation [IDA |
| Molecular Function: |
oxidized pyrimidine base lesion DNA N-glycosylase activity
[IDA DNA-(apurinic or apyrimidinic site) lyase activity [IDA oxidized purine base lesion DNA N-glycosylase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQKISKYSSM AILRKRPLVK TETGPESELL PEKRTKIKQE EVVPQPVDID WVKSLPNKQY 60
61 FEWIVVRNGN VPNRWATPLD PSILVTPAST KVPYKFQETY ARMRVLRSKI LAPVDIIGGS 120
121 SIPVTVASKC GISKEQISPR DYRLQVLLGV MLSSQTKDEV TAMAMLNIMR YCIDELHSEE 180
181 GMTLEAVLQI NETKLDELIH SVGFHTRKAK YILSTCKILQ DQFSSDVPAT INELLGLPGV 240
241 GPKMAYLTLQ KAWGKIEGIC VDVHVDRLTK LWKWVDAQKC KTPDQTRTQL QNWLPKGLWT 300
301 EINGLLVGFG QIITKSRNLG DMLQFLPPDD PRSSLDWDLQ SQLYKEIQQN IMSYPKWVKY 360
361 LEGKRELNVE AEINVKHEEK TVEETMVKLE NDISVKVED |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
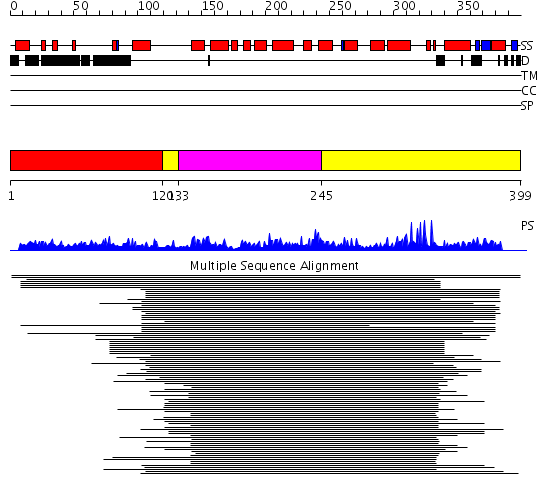
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..119] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [120..132] [245..399] |
128.751666 | Endonuclease III | |
| 3 | View Details | [133..244] | 128.751666 | Endonuclease III |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)