













| General Information: |
|
| Name(s) found: |
PRE10 /
YOR362C
[SGD]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 288 amino acids |
Gene Ontology: |
|
| Cellular Component: |
proteasome core complex, alpha-subunit complex
[IPI |
| Biological Process: |
ubiquitin-dependent protein catabolic process
[TAS |
| Molecular Function: |
endopeptidase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSIGTGYDL SNSVFSPDGR NFQVEYAVKA VENGTTSIGI KCNDGVVFAV EKLITSKLLV 60
61 PQKNVKIQVV DRHIGCVYSG LIPDGRHLVN RGREEAASFK KLYKTPIPIP AFADRLGQYV 120
121 QAHTLYNSVR PFGVSTIFGG VDKNGAHLYM LEPSGSYWGY KGAATGKGRQ SAKAELEKLV 180
181 DHHPEGLSAR EAVKQAAKII YLAHEDNKEK DFELEISWCS LSETNGLHKF VKGDLLQEAI 240
241 DFAQKEINGD DDEDEDDSDN VMSSDDENAP VATNANATTD QEGDIHLE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
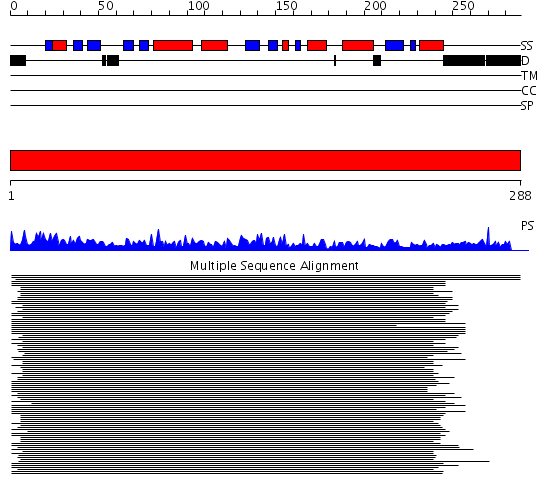
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..288] | 1703.39794 | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST IN COMPLEX WITH THE PROTEASOME ACTIVATOR PA26 FROM TRYPANOSOME BRUCEI AT 3.2 ANGSTROMS RESOLUTION |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)