













| General Information: |
|
| Name(s) found: |
SEC18 /
YBR080C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 758 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA extrinsic to plasma membrane [IDA |
| Biological Process: |
vacuole inheritance
[IDA ER to Golgi vesicle-mediated transport [TAS vacuole fusion, non-autophagic [IMP |
| Molecular Function: |
ATPase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFKIPGFGKA AANHTPPDMT NMDTRTRHLK VSNCPNNSYA LANVAAVSPN DFPNNIYIII 60
61 DNLFVFTTRH SNDIPPGTIG FNGNQRTWGG WSLNQDVQAK AFDLFKYSGK QSYLGSIDID 120
121 ISFRARGKAV STVFDQDELA KQFVRCYESQ IFSPTQYLIM EFQGHFFDLK IRNVQAIDLG 180
181 DIEPTSAVAT GIETKGILTK QTQINFFKGR DGLVNLKSSN SLRPRSNAVI RPDFKFEDLG 240
241 VGGLDKEFTK IFRRAFASRI FPPSVIEKLG ISHVKGLLLY GPPGTGKTLI ARKIGTMLNA 300
301 KEPKIVNGPE ILSKYVGSSE ENIRNLFKDA EAEYRAKGEE SSLHIIIFDE LDSVFKQRGS 360
361 RGDGTGVGDN VVNQLLAKMD GVDQLNNILV IGMTNRKDLI DSALLRPGRF EVQVEIHLPD 420
421 EKGRLQIFDI QTKKMRENNM MSDDVNLAEL AALTKNFSGA EIEGLVKSAS SFAINKTVNI 480
481 GKGATKLNTK DIAKLKVTRE DFLNALNDVT PAFGISEEDL KTCVEGGMML YSERVNSILK 540
541 NGARYVRQVR ESDKSRLVSL LIHGPAGSGK TALAAEIALK SGFPFIRLIS PNELSGMSES 600
601 AKIAYIDNTF RDAYKSPLNI LVIDSLETLV DWVPIGPRFS NNILQMLKVA LKRKPPQDRR 660
661 LLIMTTTSAY SVLQQMDILS CFDNEIAVPN MTNLDELNNV MIESNFLDDA GRVKVINELS 720
721 RSCPNFNVGI KKTLTNIETA RHDEDPVNEL VELMTQSA |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | sample: hx4 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
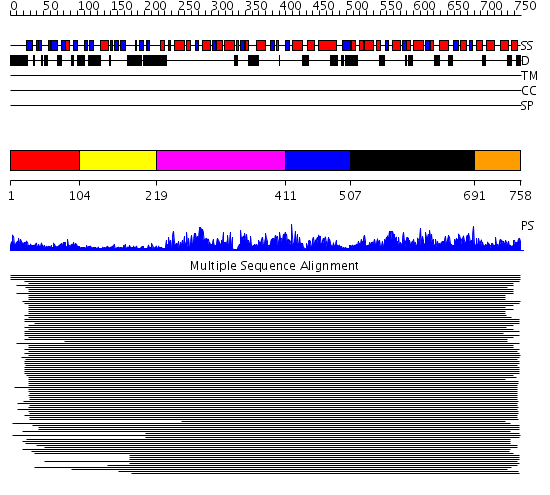
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..103] | 1136.522879 | N-terminal domain of NSF-N, NSF-Nn; C-terminal domain of NSF-N, NSF-Nc | |
| 2 | View Details | [104..218] | 1136.522879 | N-terminal domain of NSF-N, NSF-Nn; C-terminal domain of NSF-N, NSF-Nc | |
| 3 | View Details | [219..410] | 5.39794 | Holliday junction helicase RuvB | |
| 4 | View Details | [411..506] | 14.82 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 5 | View Details | [507..690] | 495.38764 | Hexamerization domain of N-ethylmalemide-sensitive fusion (NSF) protein | |
| 6 | View Details | [691..758] | 495.38764 | Hexamerization domain of N-ethylmalemide-sensitive fusion (NSF) protein |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)