













| General Information: |
|
| Name(s) found: |
ANP1 /
YEL036C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 500 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Golgi cis cisterna
[IDA alpha-1,6-mannosyltransferase complex [IPI |
| Biological Process: |
protein amino acid N-linked glycosylation
[IDA |
| Molecular Function: |
mannosyltransferase activity
[ISS alpha-1,6-mannosyltransferase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKYNNRKLSF NPTTVSIAGT LLTVFFLTRL VLSFFSISLF QLVTFQGIFK PYVPDFKNTP 60
61 SVEFYDLRNY QGNKDGWQQG DRILFCVPLR DASEHLPMFF NHLNTMTYPH NLIDLSFLVS 120
121 DSSDNTMGVL LSNLQMAQSQ QDKSKRFGNI EIYEKDFGQI IGQSFSDRHG FGAQGPRRKL 180
181 MARARNWLGS VALKPYHSWV YWRDVDVETI PTTIMEDLMH HDKDVIVPNV WRPLPDWLGN 240
241 IQPYDLNSWK ESEGGLQLAD SLDEDAVIVE GYPEYATWRP HLAYMRDPNG NPEDEMELDG 300
301 IGGVSILAKA KVFRTGSHFP AFSFEKHAET EAFGRLSRRM NYNVIGLPHY VIWHIYEPSS 360
361 DDLKHMAWMA EEEKRKLEEE RIREFYNKIW EIGFEDVRDQ WNEERDSILK NIDSTLNNKV 420
421 TVDWSEEGDG SELVDSKGDF VSPNNQQQQQ QQQQQQQQQQ QQQQQQQLDG NPQGKPLDDN 480
481 DKNKKKHPKE VPLDFDPDRN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
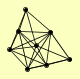
|
ANP1 FKS1 HOC1 MNN10 MNN11 MNN9 PHB1 PHB2 |
| View Details | Krogan NJ, et al. (2006) |
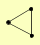
|
ANP1 PMT2 YCR043C |
| View Details | Gavin AC, et al. (2006) |
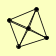
|
ANP1 HOC1 MNN10 MNN11 MNN9 |
| View Details | Gavin AC, et al. (2002) |
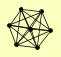
|
ANP1 FKS1 MNN10 SEC62 SEC63 SEC66 SEC72 |
| View Details | Qiu et al. (2008) |
|
ANP1 GNT1 HOC1 MNN1 MNN10 MNN11 MNN2 MNN9 OCH1 VAN1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
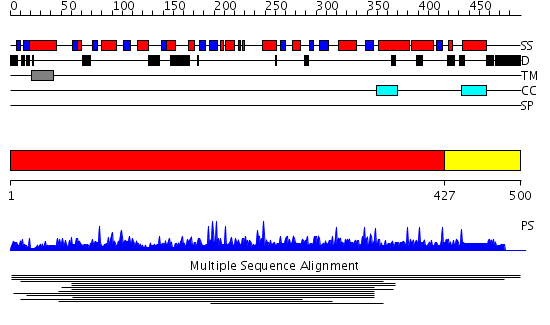
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..426] | 264.958607 | Anp1 No confident structure predictions are available. | |
| 2 | View Details | [427..500] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|