













| General Information: |
|
| Name(s) found: |
GEA1 /
YJR031C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1408 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Golgi cis cisterna
[IDA Golgi-associated vesicle [IPI |
| Biological Process: |
ER to Golgi vesicle-mediated transport
[IMP retrograde vesicle-mediated transport, Golgi to ER [IGI intra-Golgi vesicle-mediated transport [IPI actin cytoskeleton organization [IGI |
| Molecular Function: |
ARF guanyl-nucleotide exchange factor activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHDVPMETVL AVNPATMIVK ECINLCSAMN KQSRDKSQTS VAALLGGGSD IFLSQSDSFV 60
61 DSFHNLPTSS YHDPLISGLV QLRLKINDLK GLDSLNALEL LKPFLEIVSA SSVSGYTTSL 120
121 ALDSLQKVFT LKIINKTFND IQIAVRETVV ALTHCRFEAS KQISDDSVLL KVVTLLRDII 180
181 TSSFGDYLSD TIIYDVLQTT LSLACNTQRS EVLRKTAEVT IAGITVKLFT KLKLLDPPTK 240
241 TEKYINDESY TDNNLKDDII GTTTSDNDLS STDDDSAVAD DNKNEKPVQQ VIREQENDEE 300
301 TAEKAENVEP NYGITVIKDY LGLLLSLVMP ENRMKHTTSA MKLSLQLINA AIEISGDKFP 360
361 LYPRLFSLIS DPIFKSVLFI IQSSTQYSLL QATLQLFTSL VVILGDYLPM QIELTLRRIF 420
421 EILEDTTISG DVSKQKPPAI RELIIEQLSI LWIHSPAFFL QLFVNFDCNL DRSDLSIDFI 480
481 KELTKFSLPA AAVNTSNNIP PICLEGVLSL IENIYNDLQR FDRAEFVKNQ KEIDILKQRD 540
541 RKTEFILCVE TFNEKAKKGI QMLIEKGFID SDSNRDIASF LFLNNGRLNK KTIGLLLCDP 600
601 KKTSLLKEFI DLFDFKGLRV DEAIRILLTK FRLPGESQQI ERIVEAFSSK YSADQSNDKV 660
661 ELEDKKAGKN GSESMTEDDI IHVQPDADSV FVLSYSIIML NTDSHNPQVK DHMTFDDYSN 720
721 NLRGCYNGKD FPRWYLHKIY TSIKVKEIVM PEEHHGNERW FEDAWNNLIS STSVMTEMQR 780
781 DFTNPISKLA QIDILQYEKA IFSNVRDIIL KTLFKIFTVA SSDQISLRIL DAISKCTFIN 840
841 YYFSFDQSYN DTVLHLGEMT TLAQSSAKAV ELDVDSIPLV EIFVEDTGSK ISVSNQSIRL 900
901 GQNFKAQLCT VLYFQIIKEI SDPSIVSTRL WNQIVQLILK LFENLLMEPN LPFFTNFHSL 960
961 LKLPELPLPD PDISIRKAKM SRSLLSTFAS YLKGDEEPSE EDIDFSIKAF ECVKASHPLS1020
1021 SVFENNQLVS PKMIETLLSS LVIEKTSENS PYFEQELLFL LEISIILISE ASYGQEFGAL1080
1081 IADHMINISN LDGLSKEAIA RLASYKMFLV SRFDNPRDIL SDLIEHDFLV KNEIFNTKYY1140
1141 ESEWGKQVIN DLFTHLNDVK YNERALKNVK FWNFLRILIS AKDRQFAVYT FLEKYIQNGD1200
1201 IFVDDGNFMN ILSLLDEMSC AGAVGTKWEQ NYENSVEDGC EAPESNPYRS IIDLSSRSIN1260
1261 ITADLLSTVG RSNSALNKNE IIAAIQGLAH QCLNPCDELG MQALQALENI LLSRASQLRT1320
1321 EKVAVDNLLE TGLLPIFELD EIQDVKMKRI TSILSVLSKI FLGQLVEGVT SNETFLRVLN1380
1381 VFNKYVDDPT VERQLQELII SKREIEKE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
GEA1 UBP12 |
| View Details | Qiu et al. (2008) |
|
GEA1 GEA2 GLO3 SEC7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
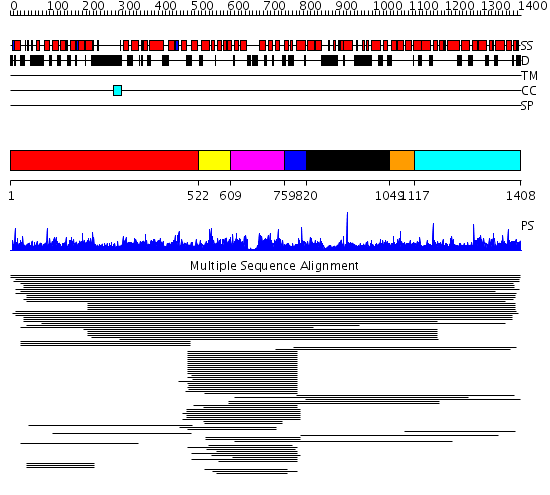
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..521] | 2.04093 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [522..608] | 918.154902 | ARF guanine-exchange factor 2, Gea2 | |
| 3 | View Details | [609..758] | 918.154902 | ARF guanine-exchange factor 2, Gea2 | |
| 4 | View Details | [759..819] | 3.39794 | Grp1 | |
| 5 | View Details | [820..1048] | 8.25 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 6 | View Details | [1049..1116] | 8.25 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 7 | View Details | [1117..1408] | 8.25 | Constant regulatory domain of protein phosphatase 2a, pr65alpha |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.79 |
Source: Reynolds et al. (2008)