













| General Information: |
|
| Name(s) found: |
TOR2 /
YKL203C
[SGD]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2474 amino acids |
Gene Ontology: |
|
| Cellular Component: |
TORC2 complex
[IPI mitochondrion [IDA extrinsic to internal side of plasma membrane [IDA vacuolar membrane [IDA TORC1 complex [IPI plasma membrane [IDA |
| Biological Process: |
cytoskeleton organization
[IMP regulation of cell growth [TAS signal transduction [TAS G1 phase of mitotic cell cycle [TAS ribosome biogenesis [IMP Rho protein signal transduction [IMP endocytosis [IMP actin filament reorganization involved in cell cycle [TAS establishment or maintenance of actin cytoskeleton polarity [IMP |
| Molecular Function: |
protein binding
[IPI protein serine/threonine kinase activity [IDA 1-phosphatidylinositol-3-kinase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKYINKYTT PPNLLSLRQR AEGKHRTRKK LTHKSHSHDD EMSTTSNTDS NHNGPNDSGR 60
61 VITGSAGHIG KISFVDSELD TTFSTLNLIF DKLKSDVPQE RASGANELST TLTSLAREVS 120
121 AEQFQRFSNS LNNKIFELIH GFTSSEKIGG ILAVDTLISF YLSTEELPNQ TSRLANYLRV 180
181 LIPSSDIEVM RLAANTLGRL TVPGGTLTSD FVEFEVRTCI DWLTLTADNN SSSSKLEYRR 240
241 HAALLIIKAL ADNSPYLLYP YVNSILDNIW VPLRDAKLII RLDAAVALGK CLTIIQDRDP 300
301 ALGKQWFQRL FQGCTHGLSL NTNDSVHATL LVFRELLSLK APYLRDKYDD IYKSTMKYKE 360
361 YKFDVIRREV YAILPLLAAF DPAIFTKKYL DRIMVHYLRY LKNIDMNAAN NSDKPFILVS 420
421 IGDIAFEVGS SISPYMTLIL DNIREGLRTK FKVRKQFEKD LFYCIGKLAC ALGPAFAKHL 480
481 NKDLLNLMLN CPMSDHMQET LMILNEKIPS LESTVNSRIL NLLSISLSGE KFIQSNQYDF 540
541 NNQFSIEKAR KSRNQSFMKK TGESNDDITD AQILIQCFKM LQLIHHQYSL TEFVRLITIS 600
601 YIEHEDSSVR KLAALTSCDL FIKDDICKQT SVHALHSVSE VLSKLLMIAI TDPVAEIRLE 660
661 ILQHLGSNFD PQLAQPDNLR LLFMALNDEI FGIQLEAIKI IGRLSSVNPA YVVPSLRKTL 720
721 LELLTQLKFS NMPKKKEESA TLLCTLINSS DEVAKPYIDP ILDVILPKCQ DASSAVASTA 780
781 LKVLGELSVV GGKEMTRYLK ELMPLIINTF QDQSNSFKRD AALTTLGQLA ASSGYVVGPL 840
841 LDYPELLGIL INILKTENNP HIRRGTVRLI GILGALDPYK HREIEVTSNS KSSVEQNAPS 900
901 IDIALLMQGV SPSNDEYYPT VVIHNLMKIL NDPSLSIHHT AAIQAIMHIF QNLGLRCVSF 960
961 LDQIIPGIIL VMRSCPPSQL DFYFQQLGSL ISIVKQHIRP HVEKIYGVIR EFFPIIKLQI1020
1021 TIISVIESIS KALEGEFKRF VPETLTFFLD ILENDQSNKR IVPIRILKSL VTFGPNLEDY1080
1081 SHLIMPIVVR MTEYSAGSLK KISIITLGRL AKNINLSEMS SRIVQALVRI LNNGDRELTK1140
1141 ATMNTLSLLL LQLGTDFVVF VPVINKALLR NRIQHSVYDQ LVNKLLNNEC LPTNIIFDKE1200
1201 NEVPERKNYE DEMQVTKLPV NQNILKNAWY CSQQKTKEDW QEWIRRLSIQ LLKESPSACL1260
1261 RSCSSLVSVY YPLARELFNA SFSSCWVELQ TSYQEDLIQA LCKALSSSEN PPEIYQMLLN1320
1321 LVEFMEHDDK PLPIPIHTLG KYAQKCHAFA KALHYKEVEF LEEPKNSTIE ALISINNQLH1380
1381 QTDSAIGILK HAQQHNELQL KETWYEKLQR WEDALAAYNE KEAAGEDSVE VMMGKLRSLY1440
1441 ALGEWEELSK LASEKWGTAK PEVKKAMAPL AAGAAWGLEQ WDEIAQYTSV MKSQSPDKEF1500
1501 YDAILCLHRN NFKKAEVHIF NARDLLVTEL SALVNESYNR AYNVVVRAQI IAELEEIIKY1560
1561 KKLPQNSDKR LTMRETWNTR LLGCQKNIDV WQRILRVRSL VIKPKEDAQV RIKFANLCRK1620
1621 SGRMALAKKV LNTLLEETDD PDHPNTAKAS PPVVYAQLKY LWATGLQDEA LKQLINFTSR1680
1681 MAHDLGLDPN NMIAQSVPQQ SKRVPRHVED YTKLLARCFL KQGEWRVCLQ PKWRLSNPDS1740
1741 ILGSYLLATH FDNTWYKAWH NWALANFEVI SMLTSVSKKK QEGSDASSVT DINEFDNGMI1800
1801 GVNTFDAKEV HYSSNLIHRH VIPAIKGFFH SISLSESSSL QDALRLLTLW FTFGGIPEAT1860
1861 QAMHEGFNLI QIGTWLEVLP QLISRIHQPN QIVSRSLLSL LSDLGKAHPQ ALVYPLMVAI1920
1921 KSESLSRQKA ALSIIEKMRI HSPVLVDQAE LVSHELIRMA VLWHEQWYEG LDDASRQFFG1980
1981 EHNTEKMFAA LEPLYEMLKR GPETLREISF QNSFGRDLND AYEWLMNYKK SKDVSNLNQA2040
2041 WDIYYNVFRK IGKQLPQLQT LELQHVSPKL LSAHDLELAV PGTRASGGKP IVKISKFEPV2100
2101 FSVISSKQRP RKFCIKGSDG KDYKYVLKGH EDIRQDSLVM QLFGLVNTLL QNDAECFRRH2160
2161 LDIQQYPAIP LSPKSGLLGW VPNSDTFHVL IREHREAKKI PLNIEHWVML QMAPDYDNLT2220
2221 LLQKVEVFTY ALNNTEGQDL YKVLWLKSRS SETWLERRTT YTRSLAVMSM TGYILGLGDR2280
2281 HPSNLMLDRI TGKVIHIDFG DCFEAAILRE KFPEKVPFRL TRMLTYAMEV SGIEGSFRIT2340
2341 CENVMKVLRD NKGSLMAILE AFAFDPLINW GFDLPTKKIE EETGIQLPVM NANELLSNGA2400
2401 ITEEEVQRVE NEHKNAIRNA RAMLVLKRIT DKLTGNDIRR FNDLDVPEQV DKLIQQATSV2460
2461 ENLCQHYIGW CPFW |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
CSI1 CSN9 ERV46 FPR2 HUL5 IQG1 MOT1 MPT5 NAS6 NMD3 NPT1 PCI8 PEP1 PST1 PUS9 RPN1 RPN10 RPN11 RPN12 RPN13 RPN14 RPN2 RPN5 RPN6 RPN7 RPN8 RPN9 RPT1 RPT2 RPT3 RPT4 RPT5 RPT6 RRI1 RRI2 SEC18 SEM1 SKP1 SPG5 SPO77 SQT1 SWA2 TCB2 TIP41 TOD6 TOR2 TSR4 UBP6 YEN1 YKL027W YMR258C YNL022C YPR003C YPT52 |
| View Details | Qiu et al. (2008) |
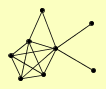
|
LSB6 LST8 MSS4 SLM1 SLM2 TOR1 TOR2 TSC11 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)